Blog Posts

clin.iobio paper
A web-based clinical diagnostic workflow to rapidly diagnose genetic disease
BY Alistair Ward ON Jan 11, 2022

clin.iobio released
Comprehensive and guided workflow to review sequencing data and report impactful variants
BY Aditya Ekawade ON Aug 25, 2020
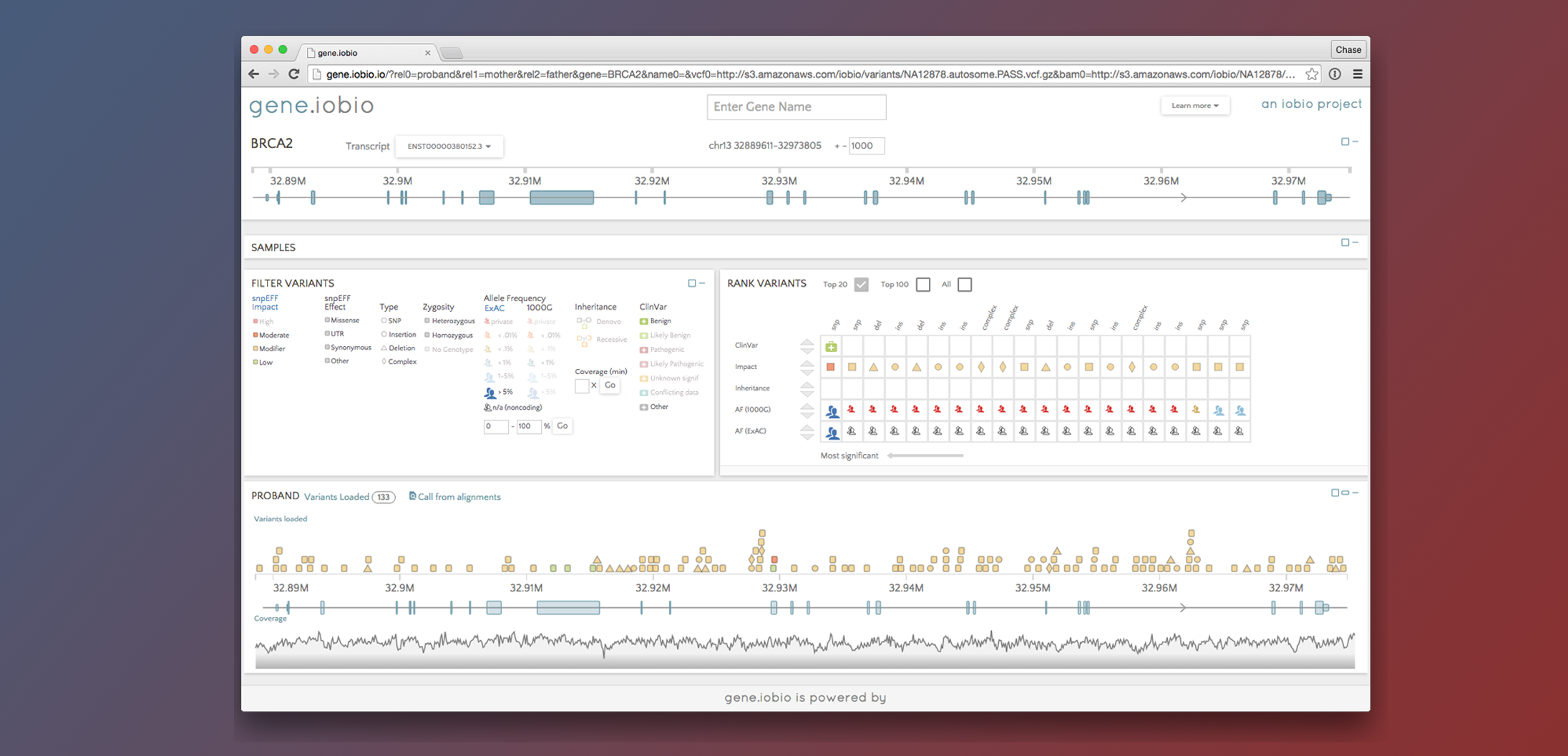
gene.iobio 4.0
Richer Variant Annotations. A Polished User Interface. Powerful New Backend
BY Tony DiSera ON Apr 20, 2020

fibridge - Access huge files in the user's browser externally over HTTP
BY Anders Pitman ON Jun 12, 2019
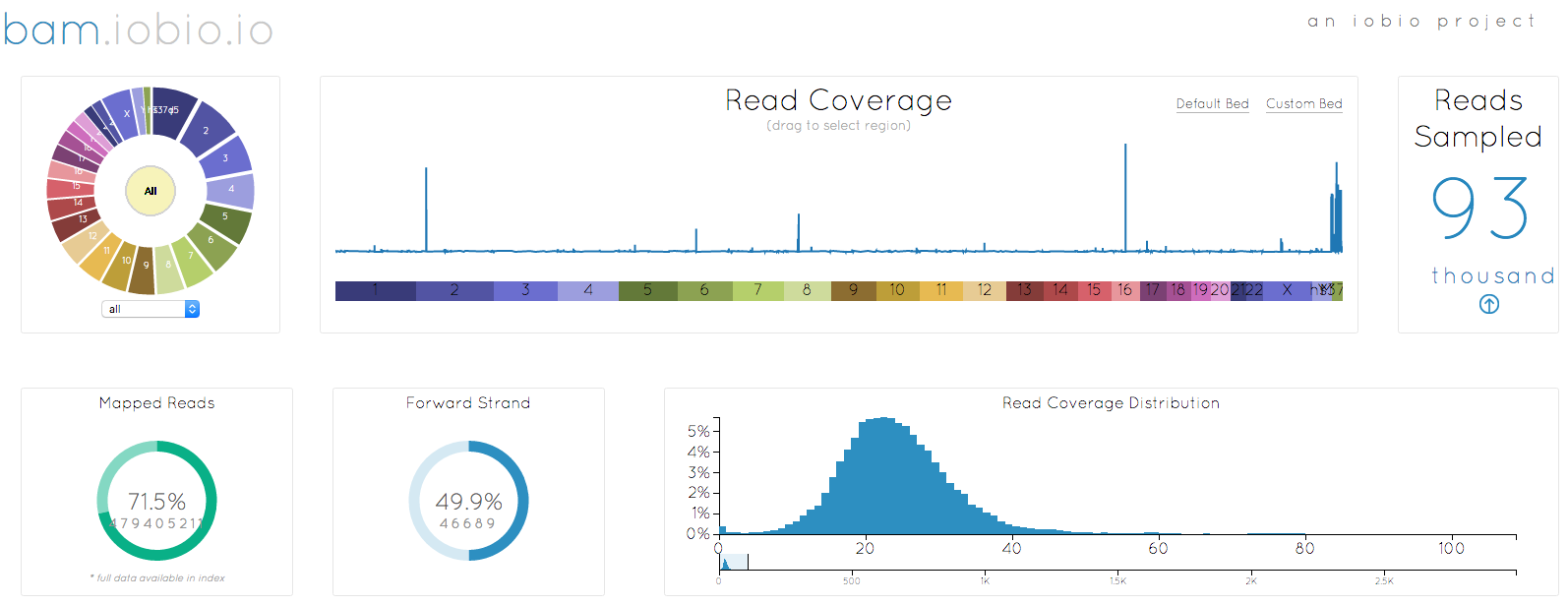
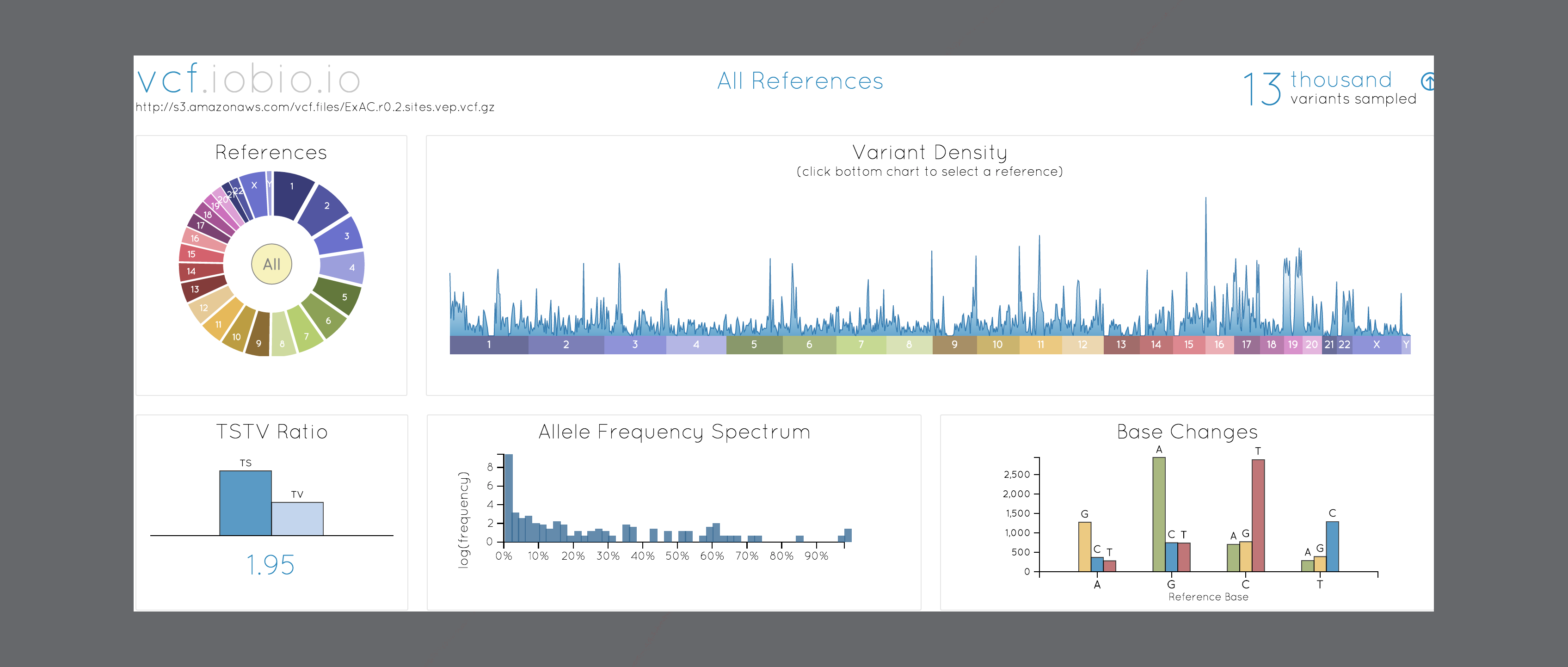
bam.iobio 2.0
Help page, local CRAM support, and improved coverage chart
BY Anders Pitman ON May 1, 2019

Genepanel.iobio Released
Generate list of genes based on suspected conditions & phenotypes
BY Aditya Ekawade ON Nov 19, 2018
ClinVar variants, gnomAD, affected status
What's new in gene.iobio 2.5.0
BY Tony DiSera ON Sep 29, 2017

Loading data into gene.iobio
Loading your variant and alignment files
BY Tony DiSera ON Sep 27, 2017

IOBIO Now in Galaxy
bam.iobio and vcf.iobio have been integrated into galaxy
BY Chase Miller ON Nov 2, 2016

Gene.iobio version 2.1
A new version of gene.iobio is now available
BY Alistair Ward ON Sep 22, 2016

Why Am I Sick? With This Tool, Doctors Could Know At a Glance
A radio interview with Gabor Marth about taxonomer.iobio
BY Gabor Marth ON Jun 15, 2016
Gene.iobio museum exhibit
Gene.iobio is featured in Genome - Unlocking Life's Code museum exhibition
BY Tony DiSera ON May 31, 2016

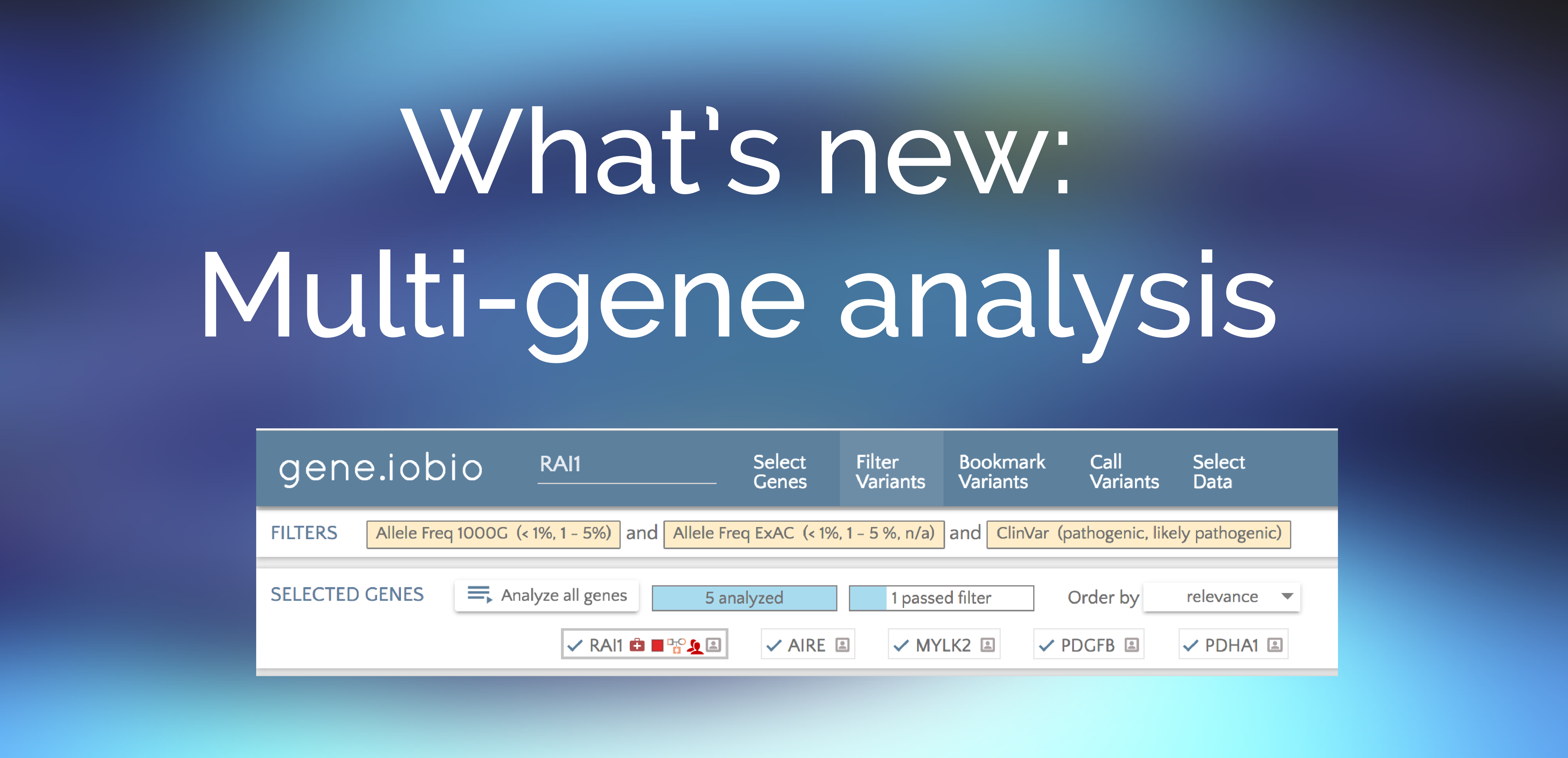
Multi gene analysis
How do I analyze lists of genes, rather than a single gene?
BY Alistair Ward ON Mar 28, 2016

What is the filter status in gene.iobio?
Only display desired variants
BY Alistair Ward ON Mar 16, 2016

Gene.iobio version 2.0
Release of version 2.0 of gene.iobio with major new functionality
BY Alistair Ward ON Mar 14, 2016
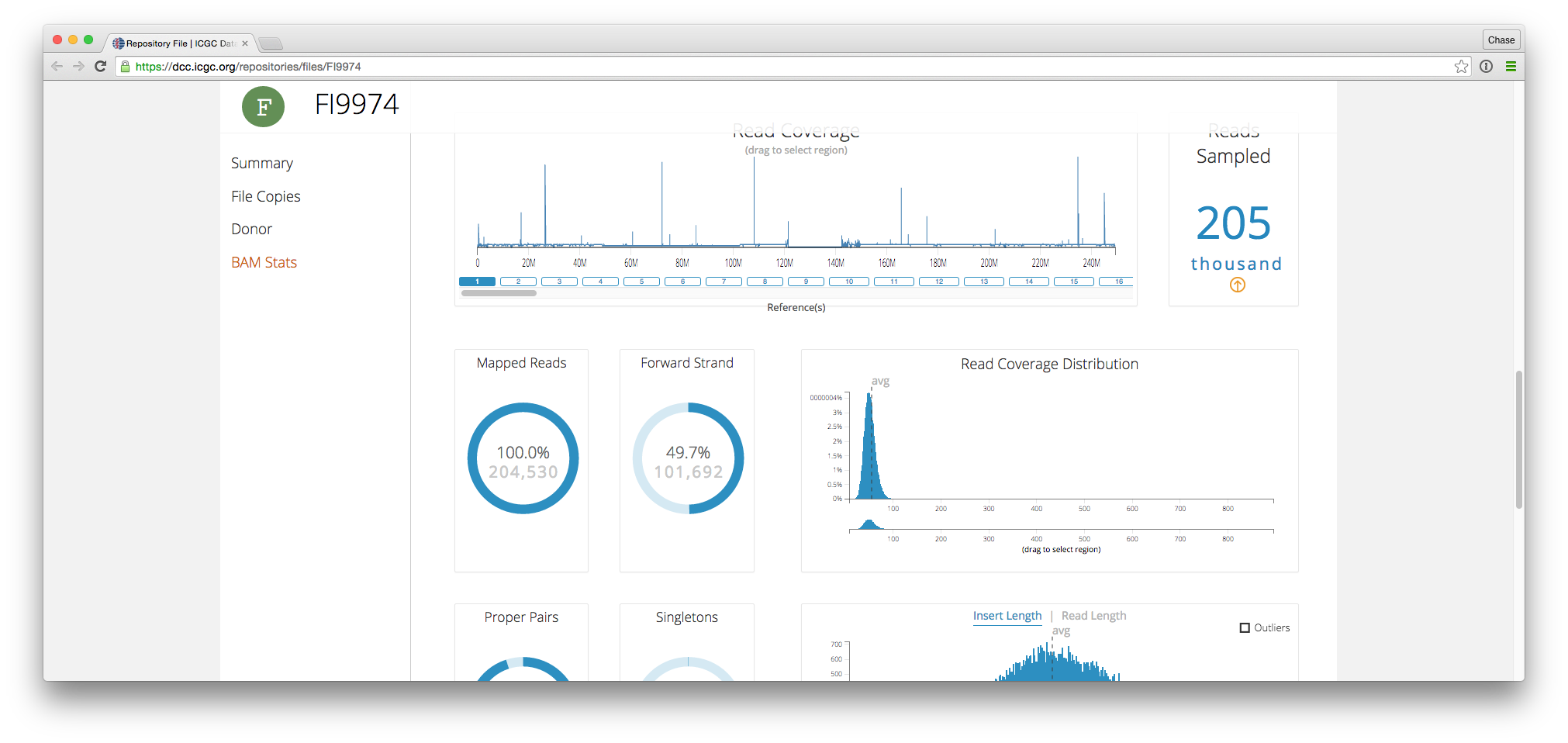
ICGC Data Portal
The ICGC data portal team integrated bam.iobio into their awesome data portal
BY Chase Miller ON Dec 8, 2015
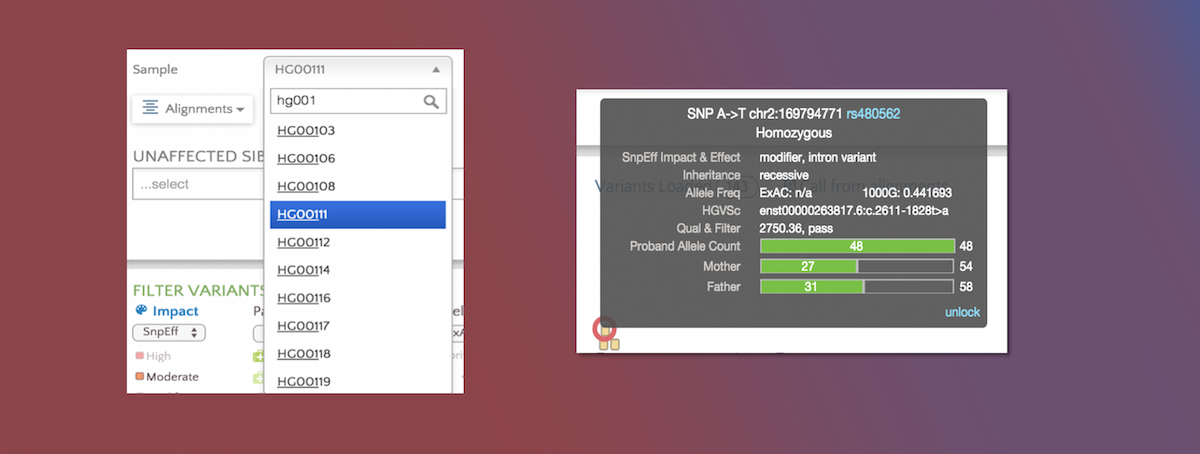
Latest Gene.iobio features
Multi-sample vcf, https support, and allele counts
BY Tony DiSera ON Sep 27, 2015

Gene.iobio integrates VEP
Polyphen & SIFT classifications, Regulatory annotations, and HGVS & dbSNP ids.
BY Tony DiSera ON Sep 7, 2015
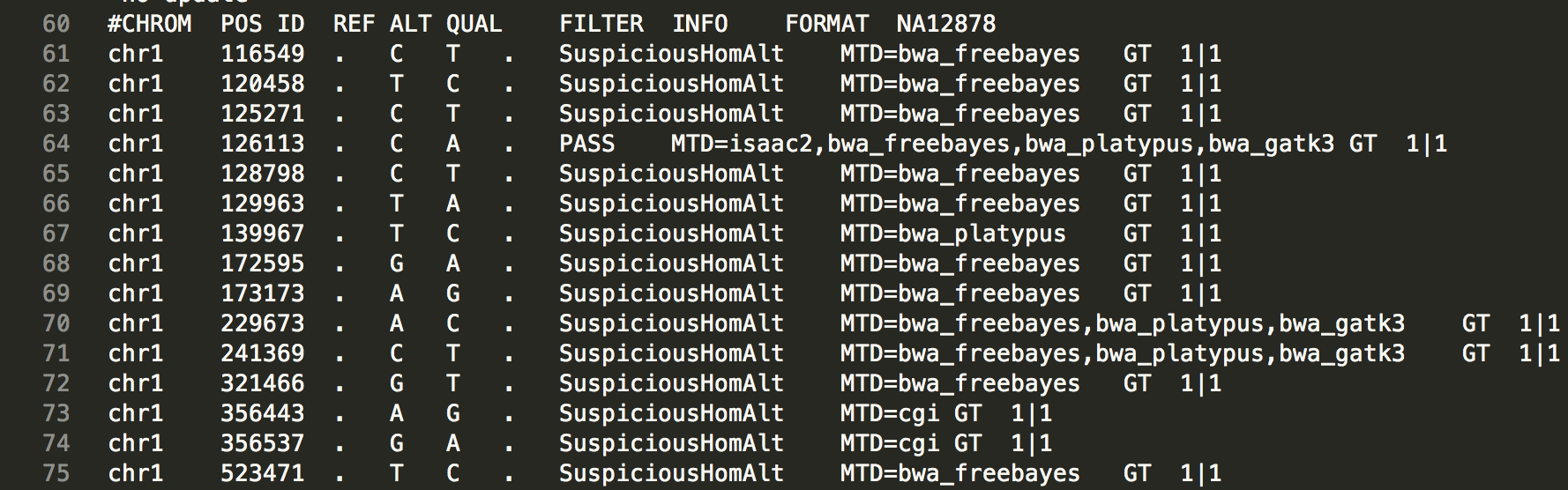
Creating a compressed and indexed VCF
The .vcf.gz and .vcf.gz.tbi file pair
BY Yi Qiao ON Sep 3, 2015

Install Locally Via Docker
We have released several docker containers to simplify the local install process
BY Yi Qiao ON Jul 5, 2015

Univeristy of Utah Blog Post on Bam.iobio
First iobio app published in Nature Methods
BY Alistair Ward ON Nov 25, 2014