As part of the analysis of a clinical dataset, we needed to answer a very simple question; is this exon important? In answering this question, we inadvertantly created a ClinVar variant viewer using gene.iobio.
The problem
When we study a child's DNA to find the genetic basis of their disease, we typically start with a list of their genetic variants and ask if any of them have been previously implicated in the disease. Then we proceed down an analysis path, looking at variants in different categories; disease-associated genes; rare variants; variants predicted to impact protein function or expression etc. and; different inheritance modes. Occasionally, we find exons with really low coverage, so no variants could be identified in them. Here is an example of a sample with no coverage in two exons in an ACMG56 gene:
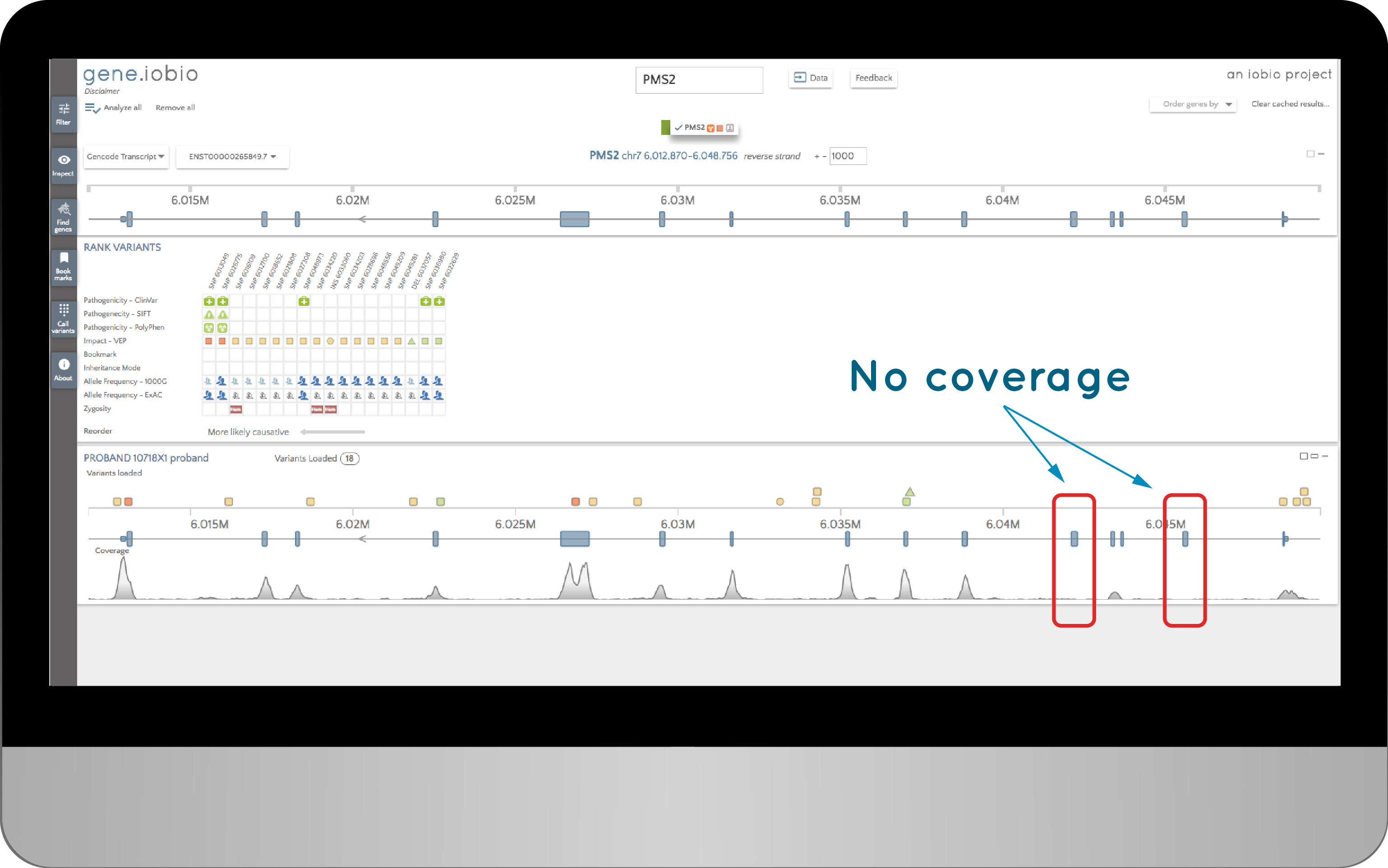
When we saw this, the question we immediately asked was, “Are these exons important?” It might be the case that these exons are predicted to exist, but are not actually present in any expressed transcript. So we needed a quick way of checking if these exons were known to harbour any interesting variants.
The solution
The solution we came up with was to ask if anybody had reported variants in these exons as being associated with a disease. One way to do this would be to look at what variants ClinVar has for this gene. So here are all the variants listed as pathogenic in ClinVar in the PMS2 gene:
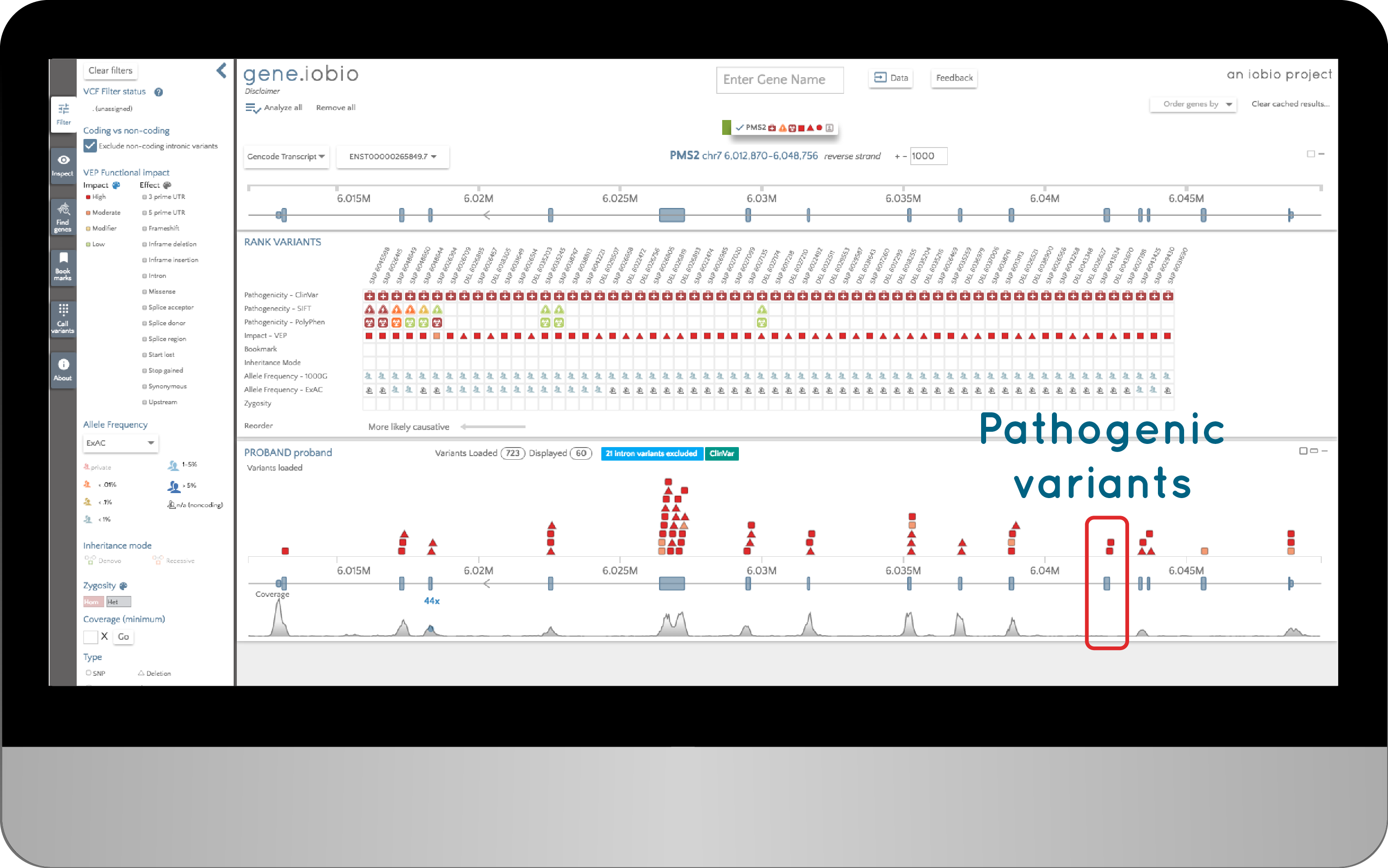
We have used the filters in gene.iobio to only show variants marked as "Pathogenic" in ClinVar. But the result is clear. The two exons with no coverage do harbour known pathogenic variants: in this case, associated with Lynch syndrome. This means that in prioritizing variants in this sample, we will inevitably have an incomplete analysis, since we know there are exons without data. We cannot rule out the possibility that this patient has pathogenic variants in these exons.
A ClinVar viewer
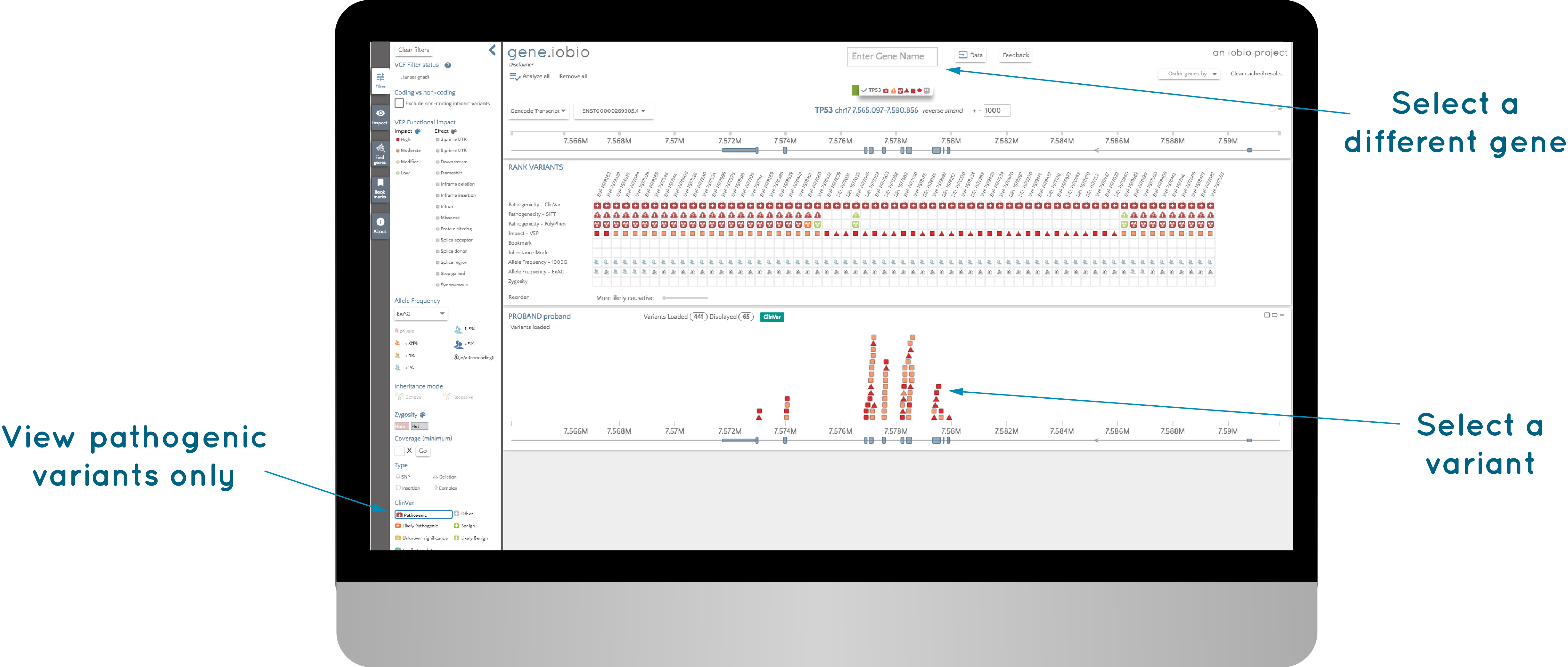
I found that this was a really useful visualization to help me understand this data set, and, realised that this was an easy way of looking at ClinVar data. Follow this link to see ClinVar variants in the TP53 gene using gene.iobio. Using the filters (at the left of the screen), we can choose to view only "Pathogenic" variants, and you can choose different genes by typing them in at the top of the page.
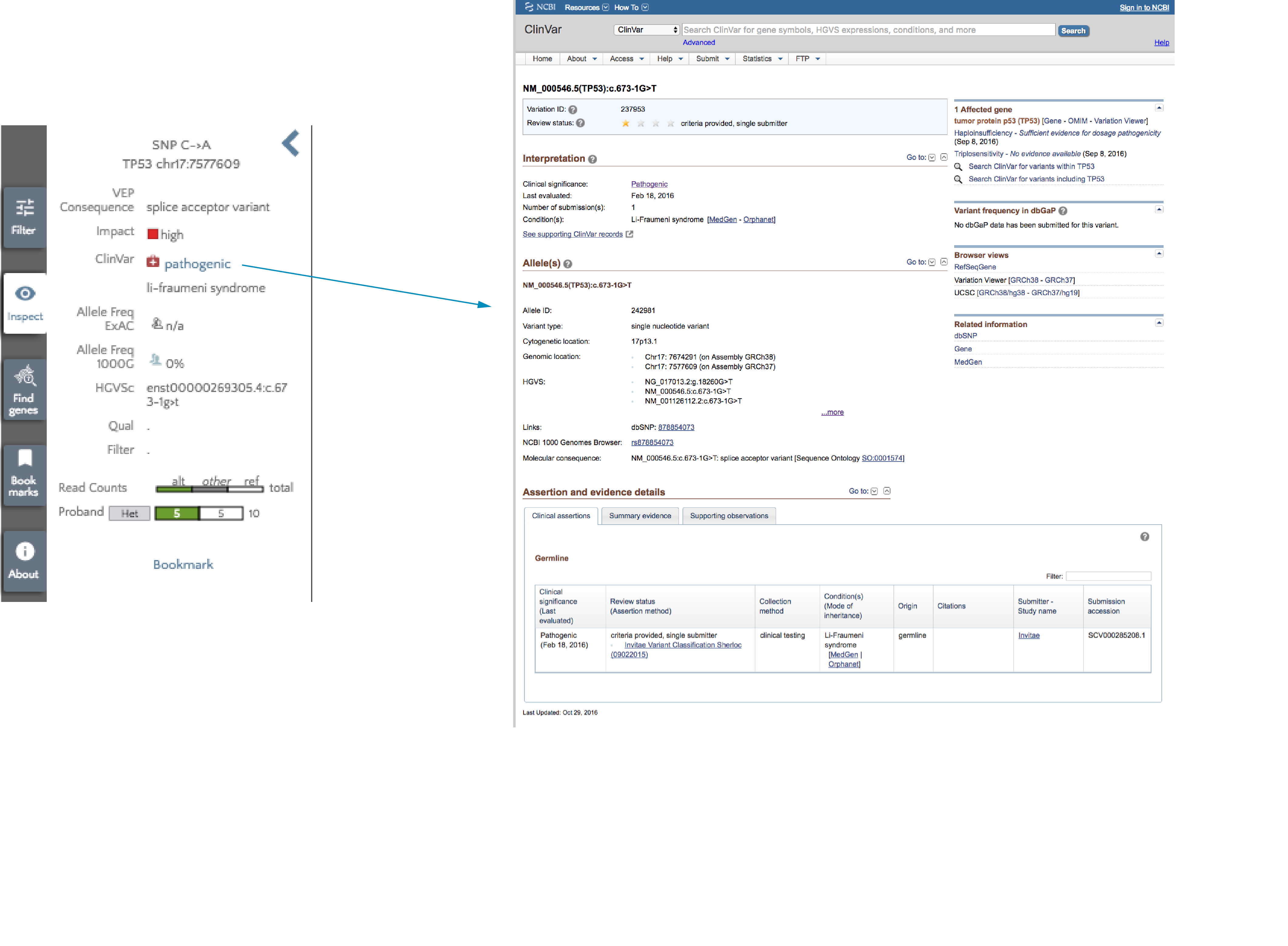
If you select any variant by clicking on it, the left panel is populated with information on the variant. The ClinVar clinical significance is listed along with any associated conditions. To get more information from ClinVar, just click the clinical significance and you will be redirected to the ClinVar page for this variant. 5