Our clin.iobio tool was recently published as part of the Journal of Personalized Medicine’s Special Issue on Precision Medicine in Clinical Practice. In this paper, we describe a real world example of using clin.iobio to rapidly diagnose genetic disease in critically ill newborns in the newborn intensive care unit (NICU).
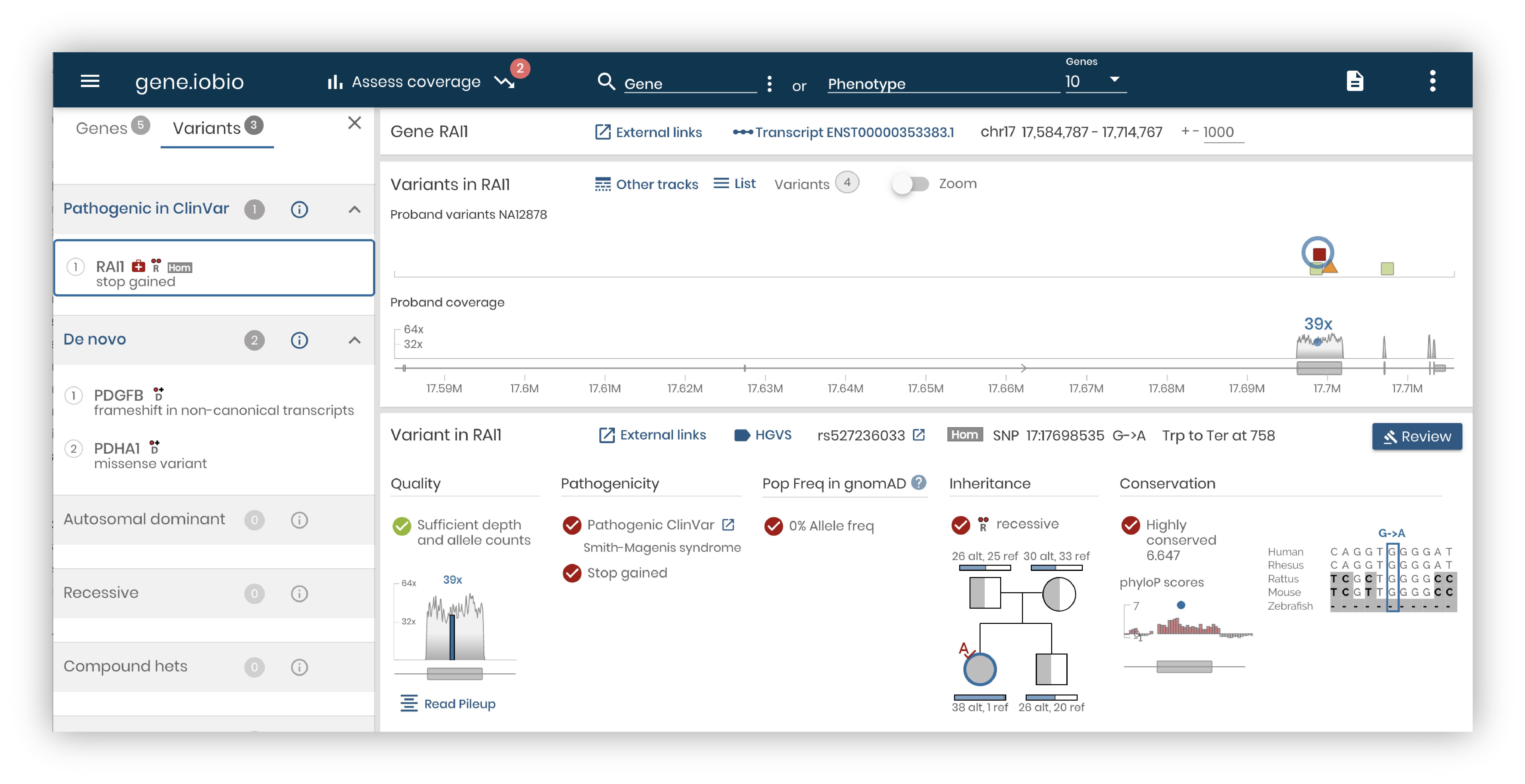
Clin.iobio is a diagnostic workflow comprising multiple steps powered by other tools in the iobio suite. In the workflow’s first step, rapid sampling of the underlying sequencing data provides an overview of basic quality metrics for the sequencing data. In the second step, our genepanel.iobio tool builds a list of candidate genes based on the provided phenotypes and/or HPO terms. These genes are then passed to gene.iobio, along with any variants prioritized by other analysis tools, for annotation, interrogation, and review. The power of gene.iobio is described in our previous blog post. Finally, a report of candidate variants flagged by the analyst are summarized in the findings page.
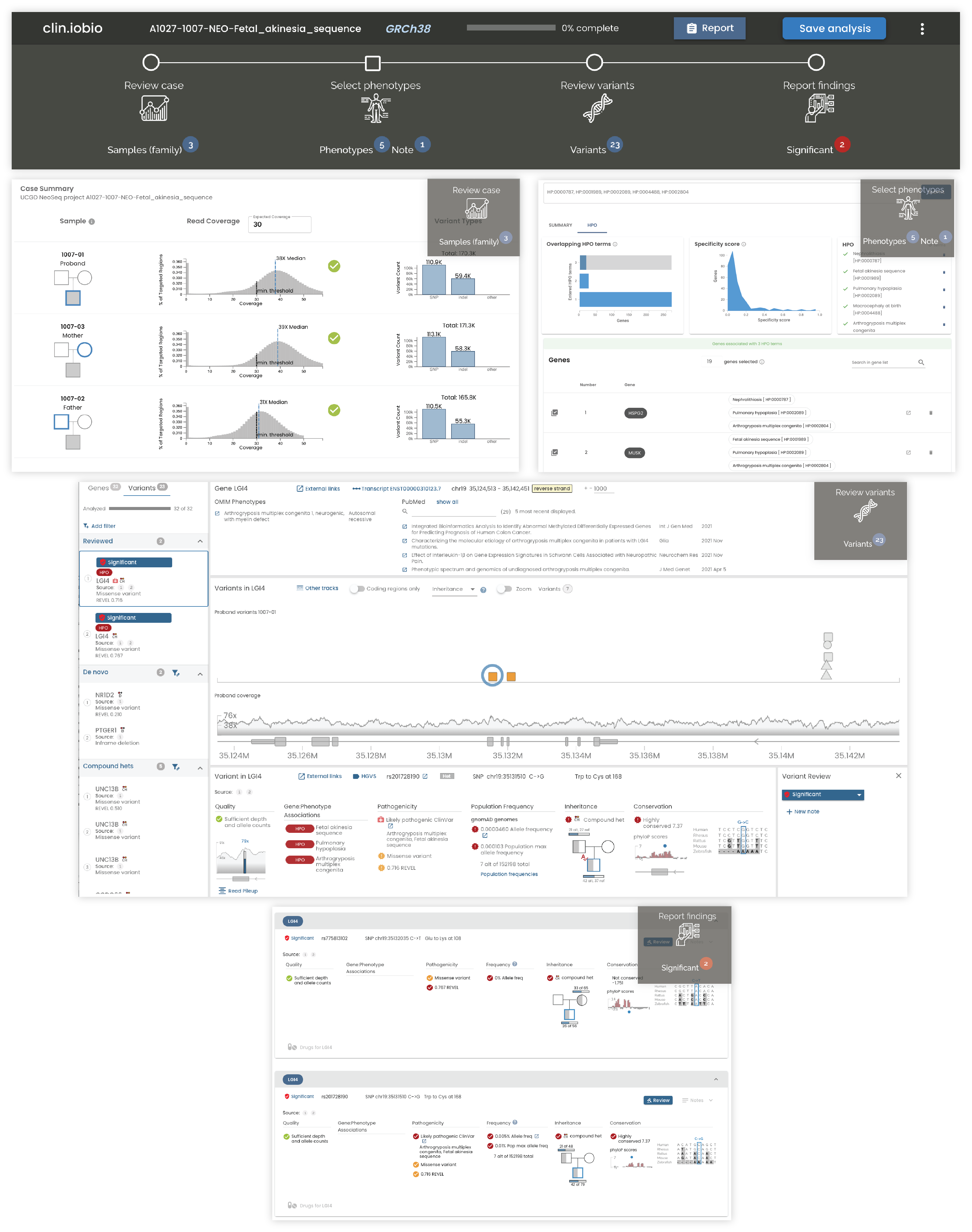
In the publication’s example, a newborn with a fetal akinesia sequence phenotype underwent rapid whole genome sequencing along with his parents. Compound heterozygous variants were quickly identified in the LGI4 gene. One of these missense variants was listed as “likely pathogenic” in ClinVar and was associated with “arthrogryposis multiplex congenita” and “fetal akinesia sequence” - the most objective phenotypes for the patient. These variants were ultimately considered diagnostic by the diagnostic team, and resulted in the newborn being moved to palliative care, sparing them from further invasive procedures.
The publication describes how clin.iobio can help all members of a diagnostic team, including those with limited computational expertise, to be actively involved in the diagnostic analysis process. This tool helps to promote a team-based approach to genomic analysis.
Please contact us at iobioproject@gmail.com with any comments or questions about gene.iobio or any of our tools in the iobio suite.