We are excited to announce our brand new app, clin.iobio! Clin.iobio provides a comprehensive and guided workflow to review sequencing data and report impactful variants. Clin.iobio also helps prioritize variants based on multiple integrated gene:disease association algorithms and knowledge bases. We hope our new comprehensive genomic analysis app will help clinical teams diagnose current and future patients!
You can try out clin.iobio right now using a demo dataset. Or you can immediately plug in your own data, by providing URLs to your BAM and VCF files.
From the clin.iobio landing page, you can import your data into the app using the provided screen prompts for entering data URLs or a configuration file (from a previous analysis). Our screencast will help guide you through these options.
Clin.iobio is also easily launchable through genomic data platforms such as Mosaic by Frameshift. Using Mosaic, a user can immediately launch a project into clin.iobio to review genes or variants.
Here are the main clin.iobio workflow steps:
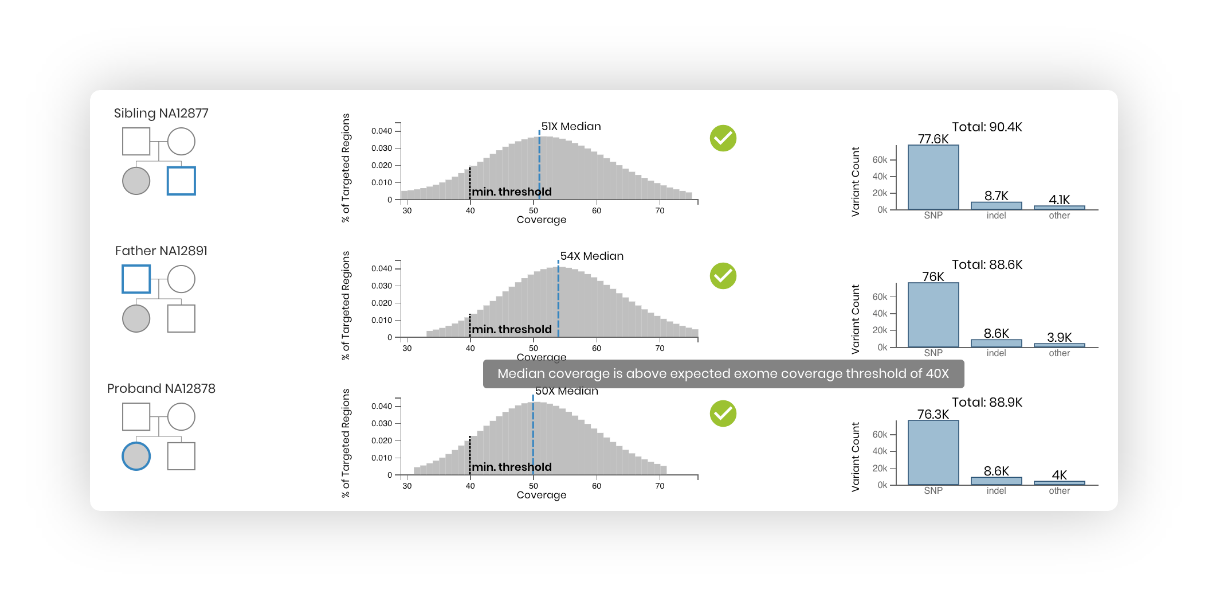
Review case:
Review relatedness, clinical notes sequencing data, and quality metrics (read coverage distribution and variant types).
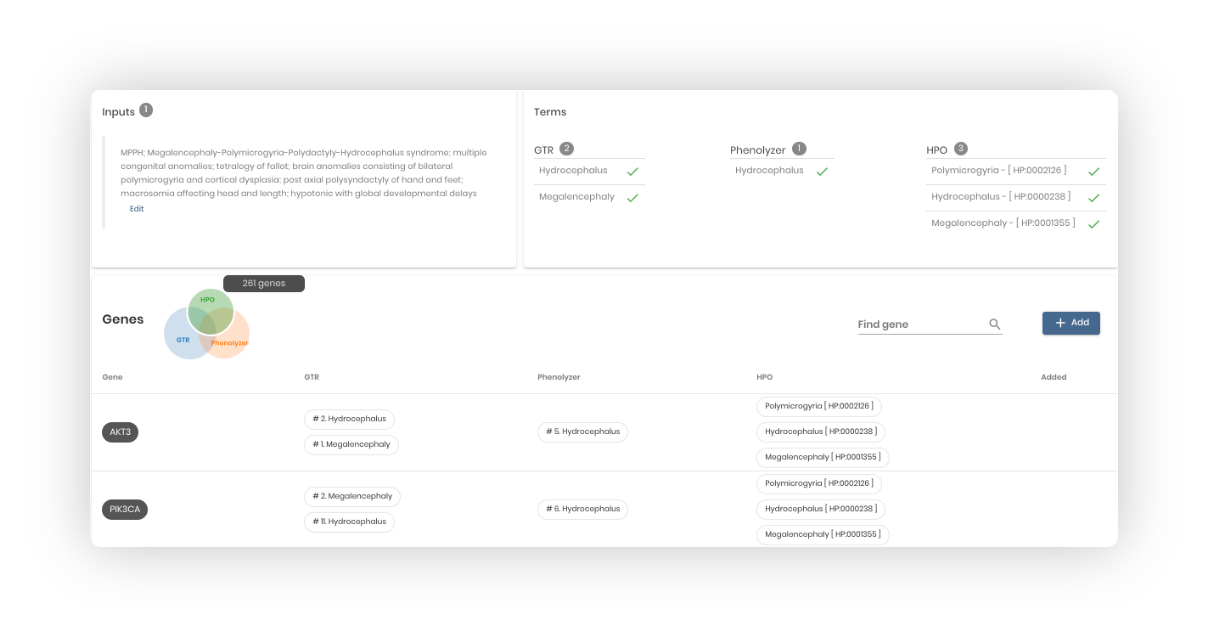
Define phenotypes:
Enter a clinical note and select the suspected genetic disorders and/or phenotypes. Terms are automatically extracted and a disease-prioritized gene list is provided for review in the next step.
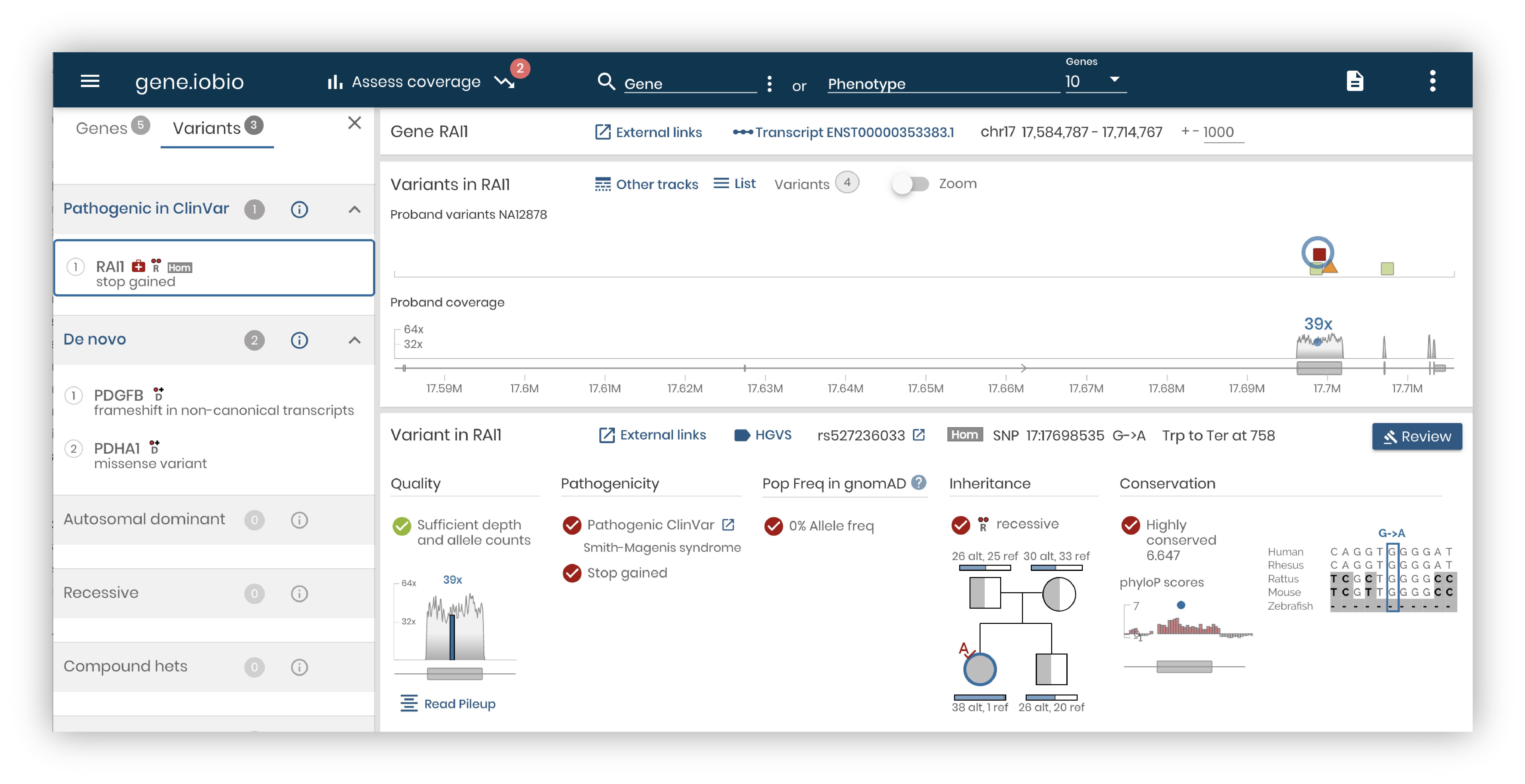
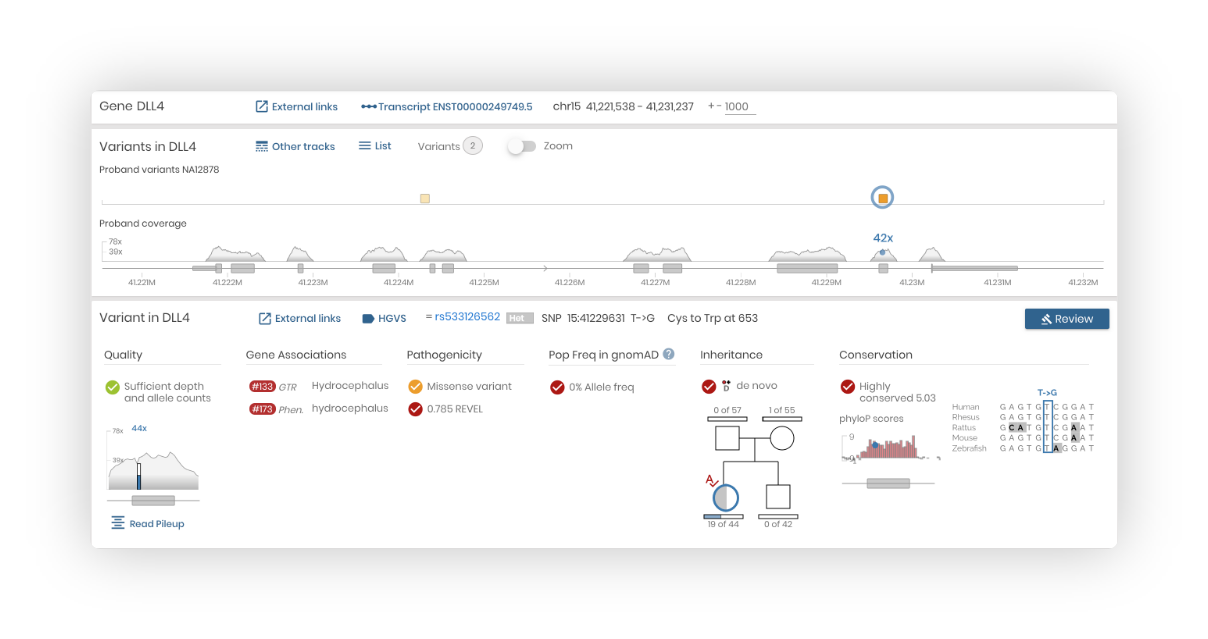
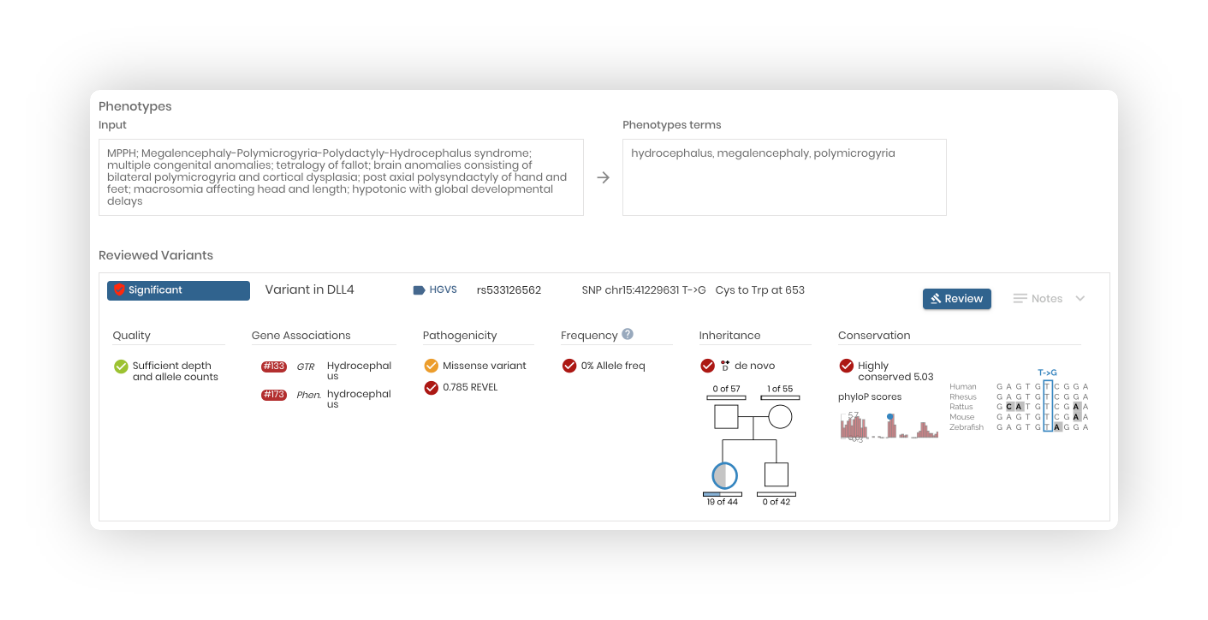
Review variants:
Review variants from your entered gene or variant list. You can review variants and attach a significance to any loaded variant.
Report findings:
You can report your findings from all aspects of the workflow, including any reviewed variants. Within the “Report Findings” step you can export a PDF report. You can also find relevant drug information for the genes in your report. You can also download a file to immediately resume your analysis at a later time.
We are very excited about our new clinical iobio app! We hope you find it useful! Please help us to improve it. Try it out and contact us at iobioproject@gmail.com with any feedback or questions!