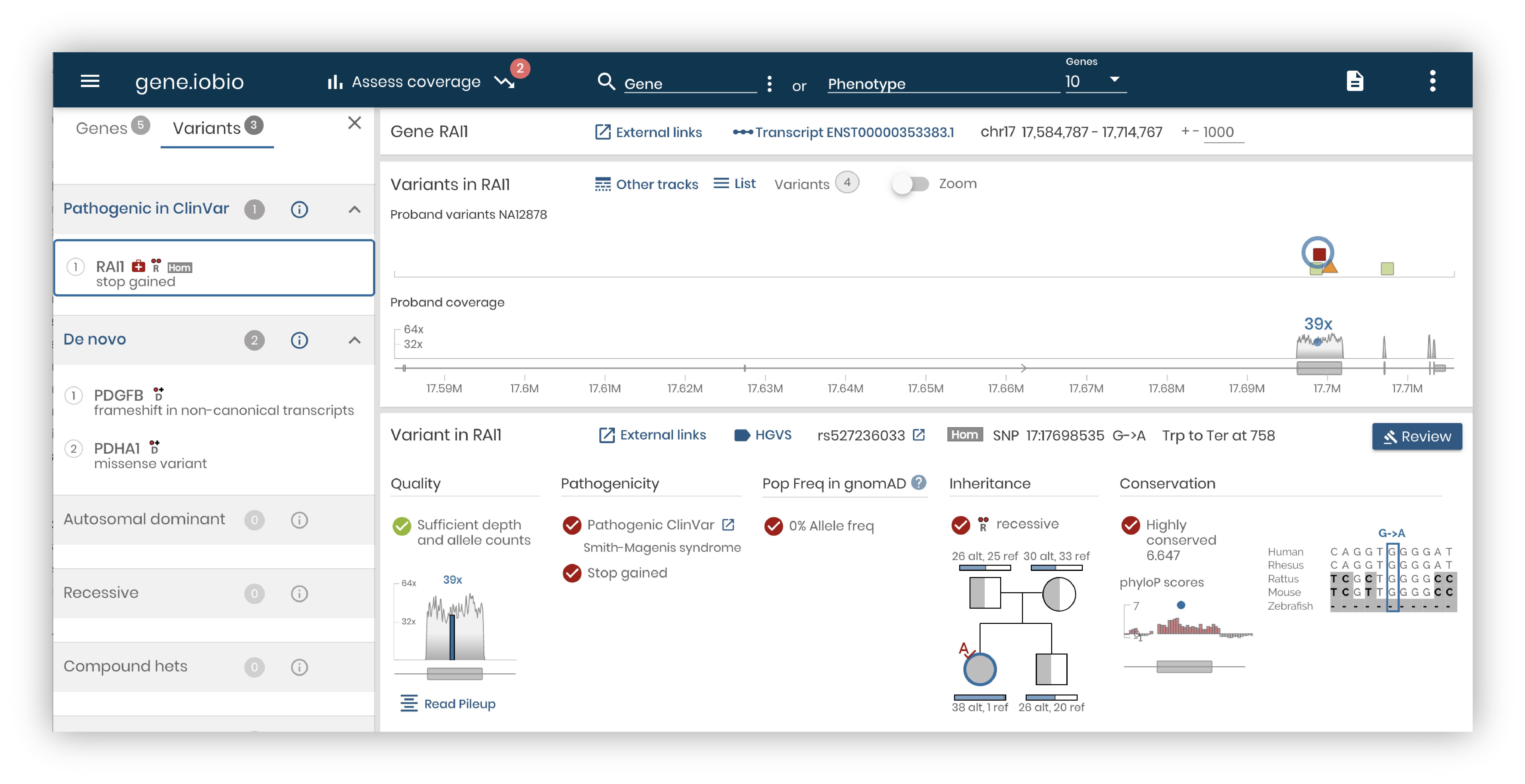
Our powerful variant interrogation and prioritization tool, gene.iobio, was recently published in Nature Scientific Reports. Our previous blog post describes gene.iobio in some detail, highlighting features that have been added as the tool has evolved. The publication is an interesting dive into how genomic data is used in clinical diagnostics, and describes specific use cases where the real-time analysis and intuitive visualization in gene.iobio support the analysis team.
For example, the case of a young boy with dysgenesis of the corpus callosum with retrocerebellar fluid collection, laryngeal cleft, among other primary objective findings and less specific phenotypes, was analysed by a clinical team.
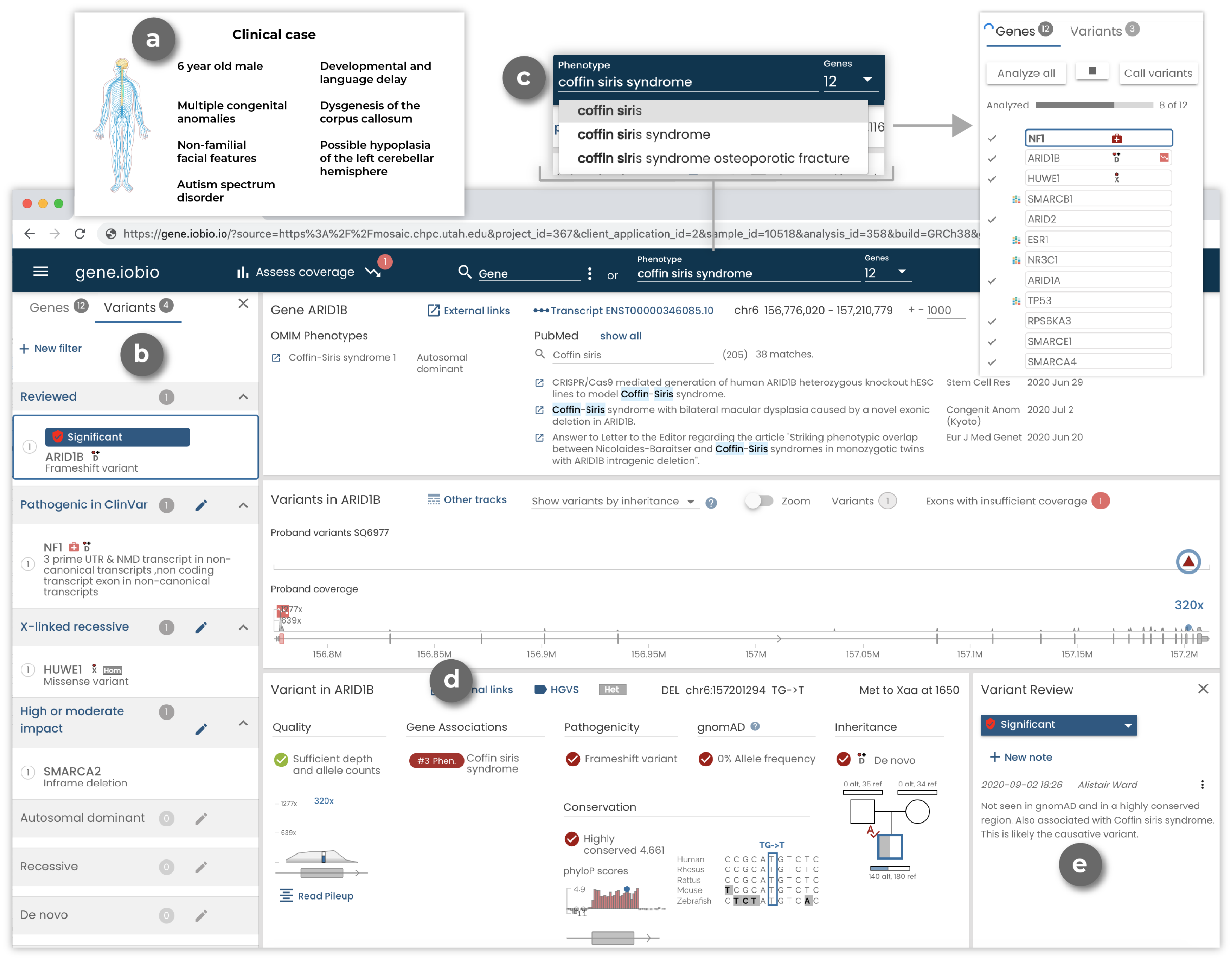
The results returned by a commercial sequencing provider (a likely pathogenic synonymous variant) were questioned by the geneticist working with the family and wanted to perform an independent review of the case. One of the candidate variants reviewed in gene.iobio was a rare de novo frameshift mutation in ARID1B. With access to up-to-date variant annotations, OMIM phenotypes, PubMed publications, and phenotype driven gene list generation, the clinical team was able to conclude that this was the causative variant for the patient.
The publication describes cases where gene.iobio was used to both refute or confirm the role of variants in patients and assess all available evidence (including sequencing coverage, false negative / positive calls etc) in an easy-to-use, visual environment. We believe the paper provides a comprehensive view of the role of sequencing, and how tools need to adapt to support the teams using this data to diagnose patients with a wide variety of diseases and phenotypes.
Please contact us at iobioproject@gmail.com with any comments or questions about gene.iobio or any of our tools in the iobio suite.