Today we’re excited to release a brand new app, genepanel.iobio, to the iobio suite. This tool will allow you to build gene lists for use in other iobio tools or in other analyses. Regardless of where you use your gene lists, genepanel.iobio strives to provide you with the most pertinent gene list possible, furthering your genetic diagnosis capabilities.
Armed with the knowledge of the NCBI’s genetic testing registry (GTR), Phenolyzer from the Wang Genomics Lab, and your own personal experience, genepanel.iobio integrates all of these sources into a modern, intuitive web app. These resources have a rich array of information, but extracting a simple list of genes can be difficult. This new app makes it easy by pulling only the most relevant information in real-time
Genepanel.iobio is open-source and free for academic use. We think it is extremely useful as a standalone app, but is especially valuable in generating lists of genes for powering variant interrogation in our gene.iobio app. You can quickly generate high confidence gene lists, based on the presence of a gene on many commercial panel tests, modes of inheritance etc.; choose to consider panels submitted to GTR by specific vendors, or review the panel tests associated with the searched conditions. These lists can be easily expanded or contracted to support your downstream research. You can learn more about how to use the GTR and Phenolyzer resources in genepanel.iobio.
This is a brand new app, and we’d love to hear your opinions. If you have any comments or suggestions, please don’t hesitate to tell us how we can improve this app!
Workflow
This simple example will demonstrate the workflow of the application. Here we show how curate a comprehensive list of genes for the condition “Dejerine-Sottas disease” (an inherited neurological disorder) based on the syndrome and associated phenotypes.
Generating a gene list using the Genetic Testing Registry
Start by typing the condition name “Dejerine-Sottas disease” in the search box provided. Search term will be autocompleted as you type. You can also enter free-form text if a term doesn’t match exactly.
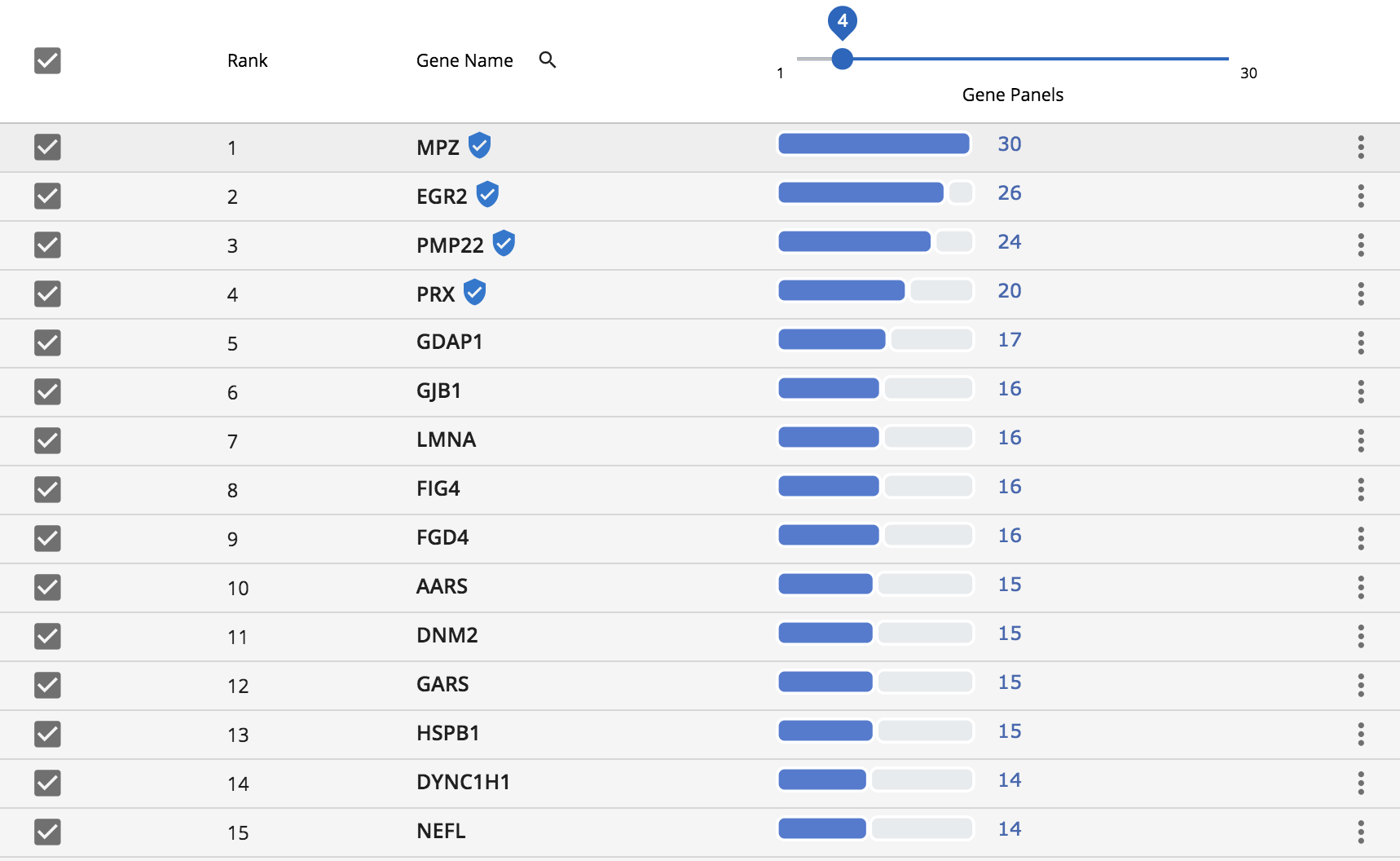
Gene results are then displayed in a simple table, with genes occurring in the most panels at the top. The genes marked with the “shield icon” are genes that GTR defines as known associated genes. Often times, mutations within these genes are what define the specific disorder. These genes are given priority and occur at the top of the list.
Generating a gene list using the Phenolyzer
Similar to the GTR, type the phenotype terms in the search box provided. In this example, we search for “pes cavus”, “distal muscle weakness” and “hammertoes” which are some of the phenotypes associated with the “Dejerine-Sottas disease”.
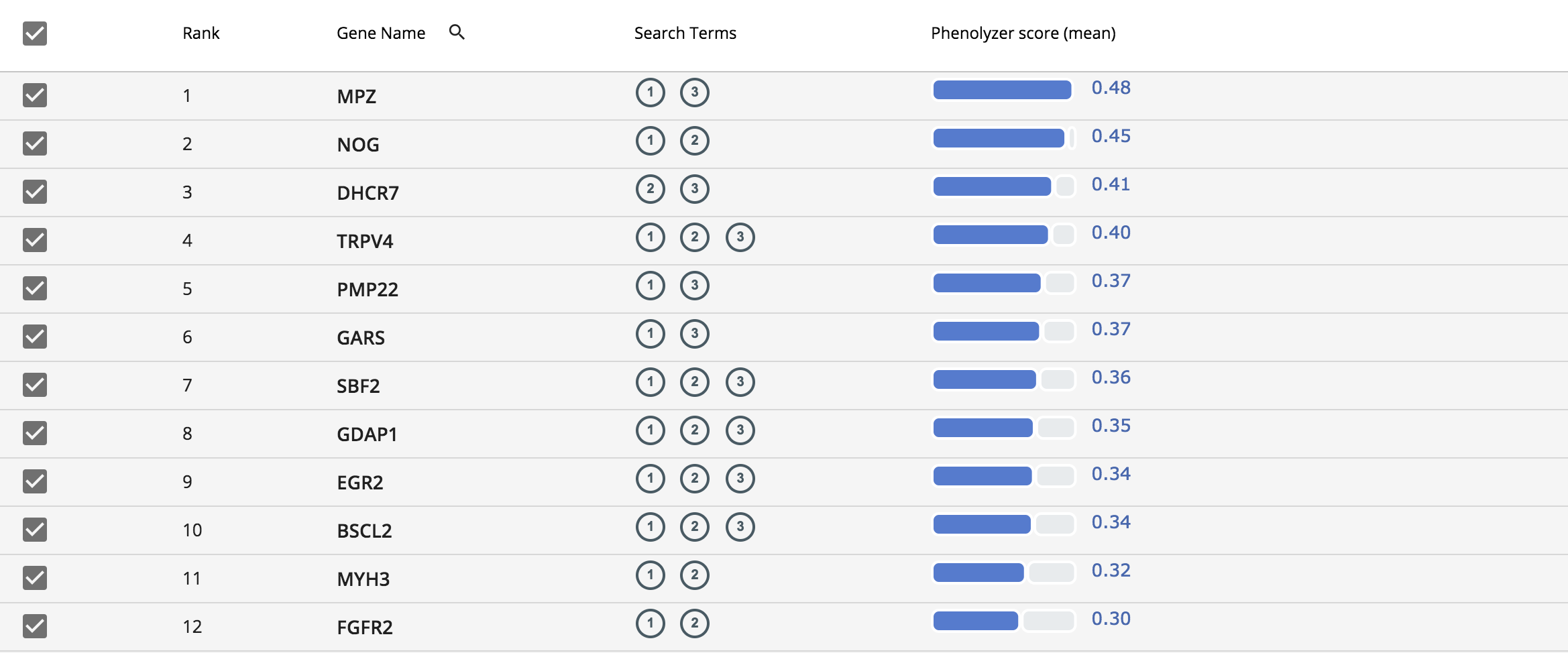
The resulting gene list is displayed in a simple table and it is sorted based on the Phenolyzer score. The column “Search Term” in the table indicates the gene associated with its respective phenotype. For example (1) in this column indicates “distal muscle weakness, (2) indicates “pes cavus” and (3) indicates “hammertoes”.
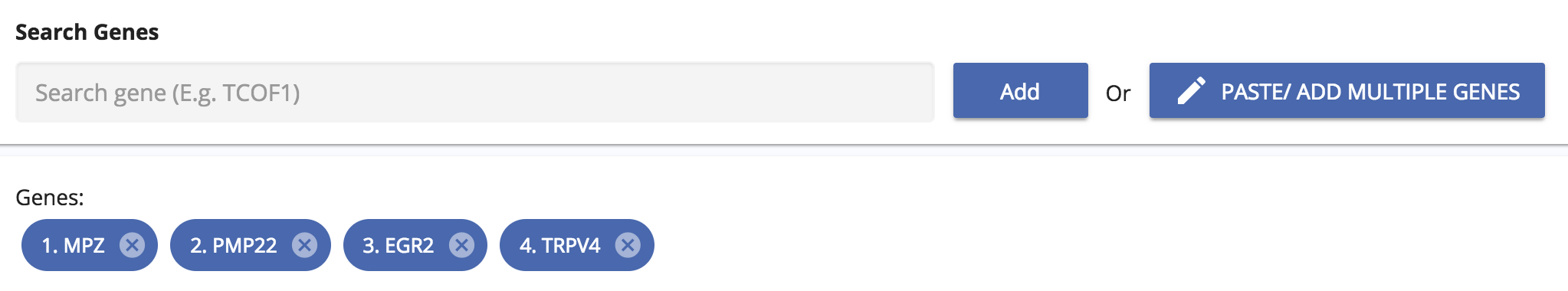
Adding your own gene list
The app also allows you to add your own gene lists. These can be simply entered into the search box or copied and pasted as multiple genes. The app will also autocomplete gene names and alert you if a gene name isn’t right.
Summarizing results from all resources
The results from all the resources are propagated to the summary page. From our example above, here is a comprehensive gene list for Dejerine-Sottas disease and its associated phenotypes, as well as some manually added genes.
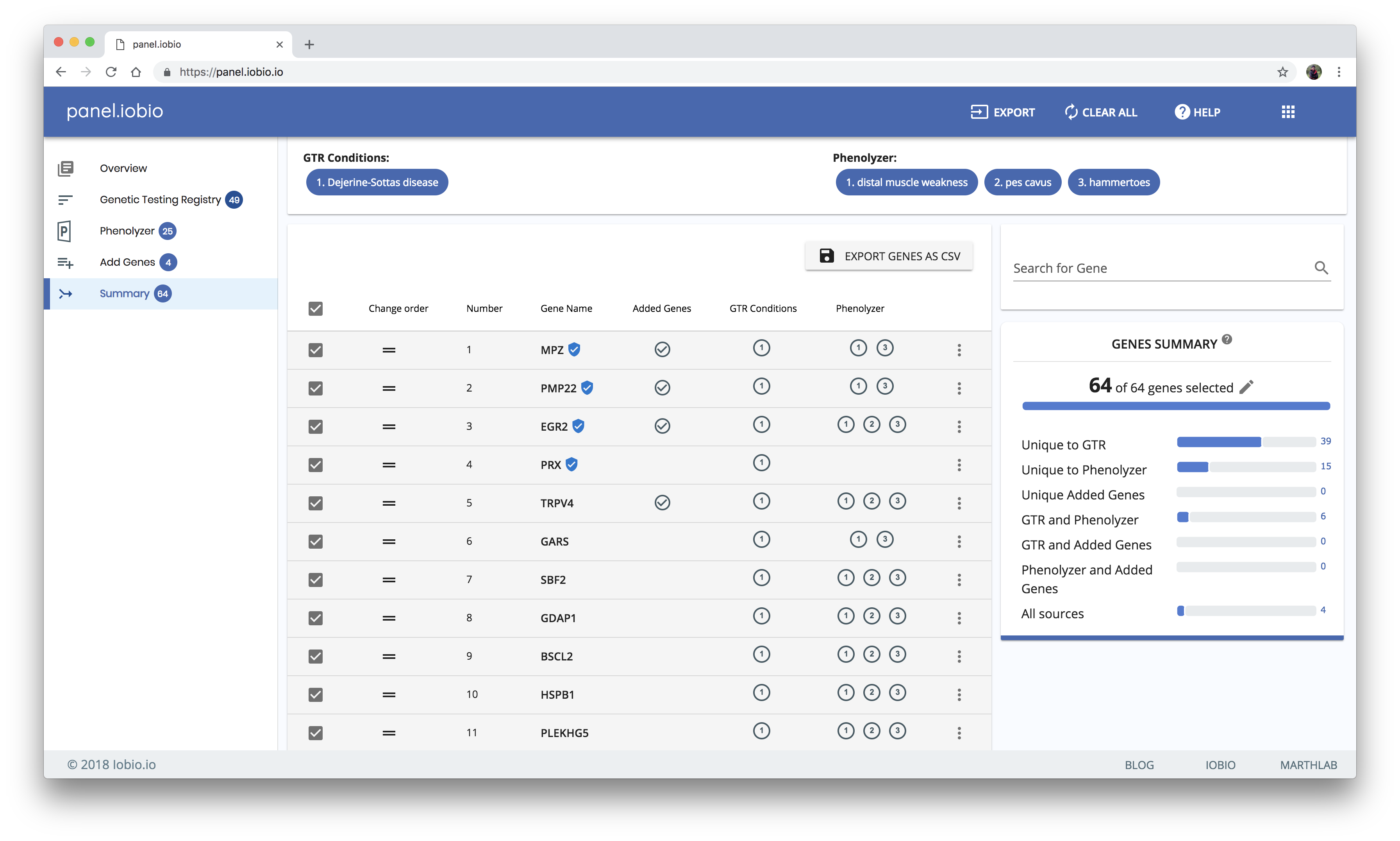
You can further refine your gene list in this summary view by checking and unchecking the boxes and dragging to reorder genes in the list. Lastly, your list can be exported by copying it to your clipboard, by saving it to a text file, or by saving it as a comma-separated value (csv) file that includes additional information for your genes.
With all of this functionality, hopefully, you can see why we’re so excited to release this new app to our suite of iobio tools!



