In March 2013, an The American College of Medical Genetics and Genomics (ACMG) working group released a list of 57 genes (later revised to 56), that harbour mutations that when identified, could lead to severe outcomes, but were medically actionable. These genes are implicated in a number of disorders, including inherited cancers, inherited cardiac diseases, connective tissue diseases affecting the cardiovascular system, familial hypercholesterolemia, and malignant hyperthermia susceptibility. The ACMG also generated a list of recommendations that state that known pathogenic (or in some cases expected pathogenic) variants discovered in any of these genes should be reported regardless of an individuals age. These recommendations referred to what are now known as secondary findings in an individual undergoing exome or genome sequencing.
It was felt that ethical concerns around testing minors for secondary findings was compensated by the potential benefit to the future health of the individual and their family. As such, the working group did not support an option to allow an opt-out of receiving this information, however, in March 2014, the recommendations were relaxed to allow an opt-out. It was then recommended that individuals undergoing exome or genome sequencing should undergo counseling before and after testing. Annual updates to the ACMG policy on secondary findings is expected as sequencing becomes more ubiquitous and our ability to interpret findings improves.
The importance of these genes is clear, and it is likely that when analyzing an individual or a family in gene.iobio, looking at variants in them will be desirable. To make this an easy task for the user, we have made the ACMG56 genes a default list that can be easily selected for analysis. If we open up gene.iobio with the modified 1000 Genomes exome trio dataset loaded, click on 'Genes' at the left of the screen, we can then select 'Import ACMG56 Genes'.
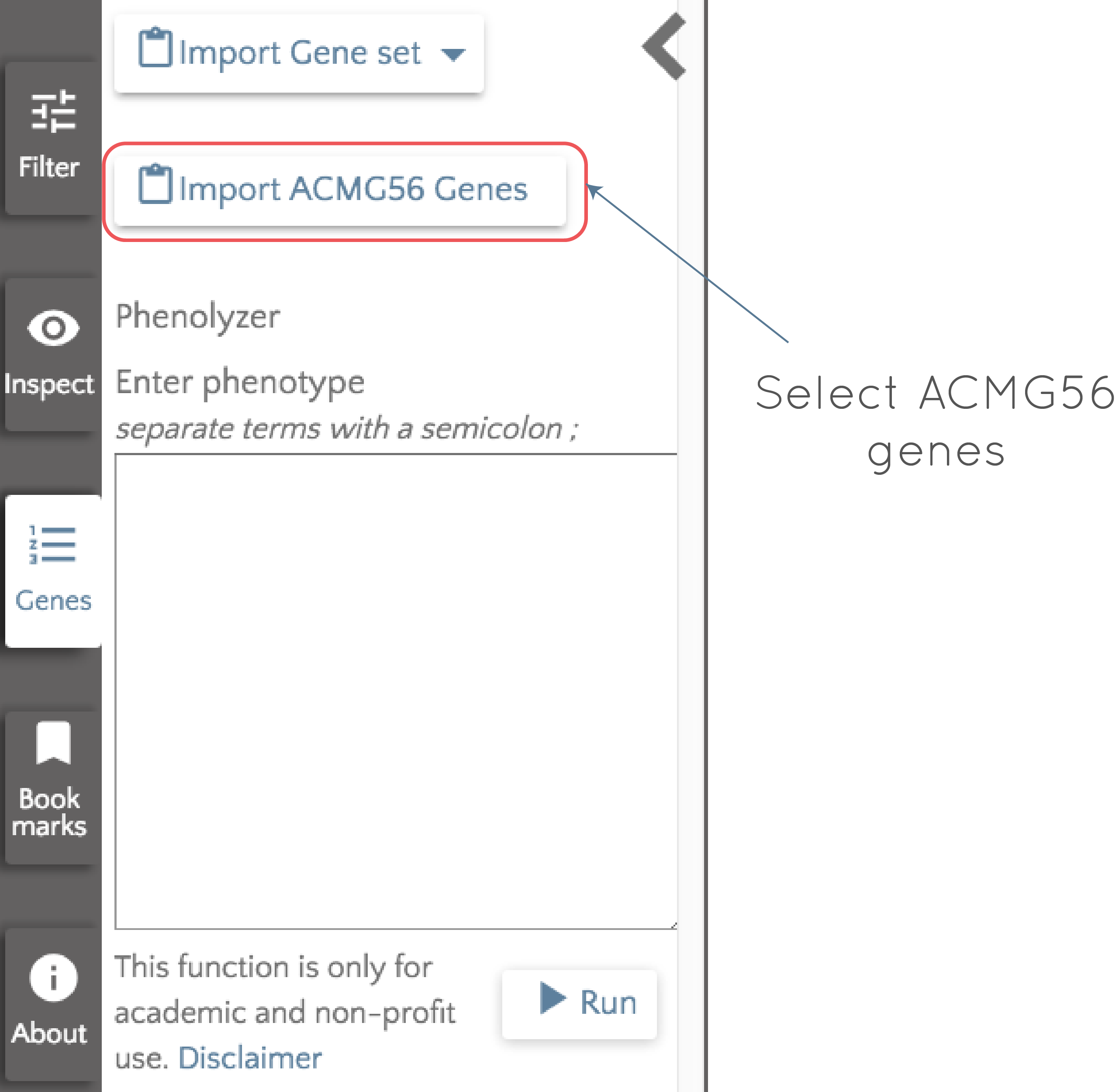
Once selected, the ACMG56 genes replace all other genes that were being analyzed and appear as gene badges (as discussed in the previous post). If we then select 'Analyze all', each of the genes is analyzed in turn. By clicking on the gene badge, we can quickly jump to any of the analyzed genes and assess the contained variants, while processing continues through the list. If we perform the analysis on the ACMG56 genes for this modified exome trio dataset, we see the following:
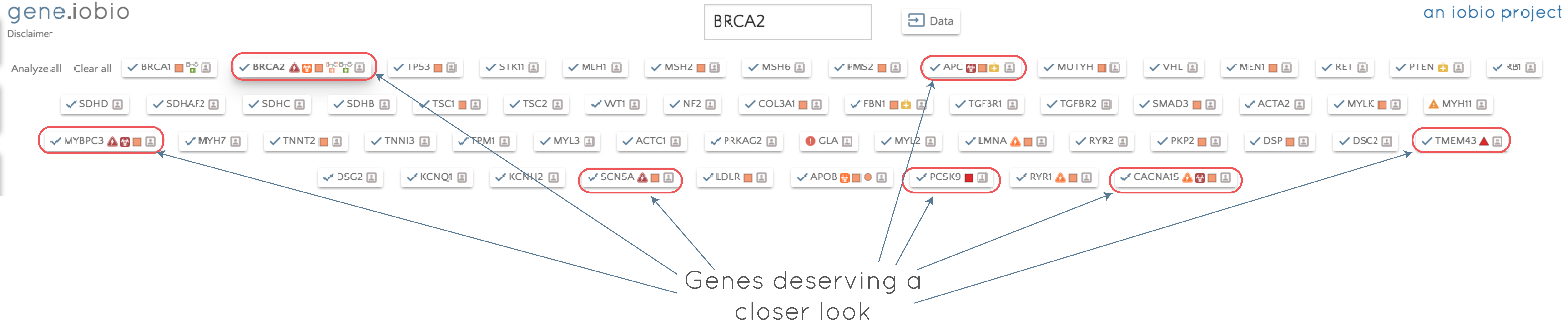
With almost no effort on our part, we have scanned for secondary findings in this individual. The results suggest that we should take a closer look at BRCA2, APC, MYBPC3, TMEM43, SCN5A, PCSK9 and CACNA1S. If we take a look in the APC gene, for example, we can see that the child has a heterozygous missense variant inherited from her mother that is listed as of uncertain significance in ClinVar, but is predicted to be possibly_damaging by PolyPhen. The variant has very low allele frequency in the 1000 Genomes data (0.02%) and is not seen in ExAC.
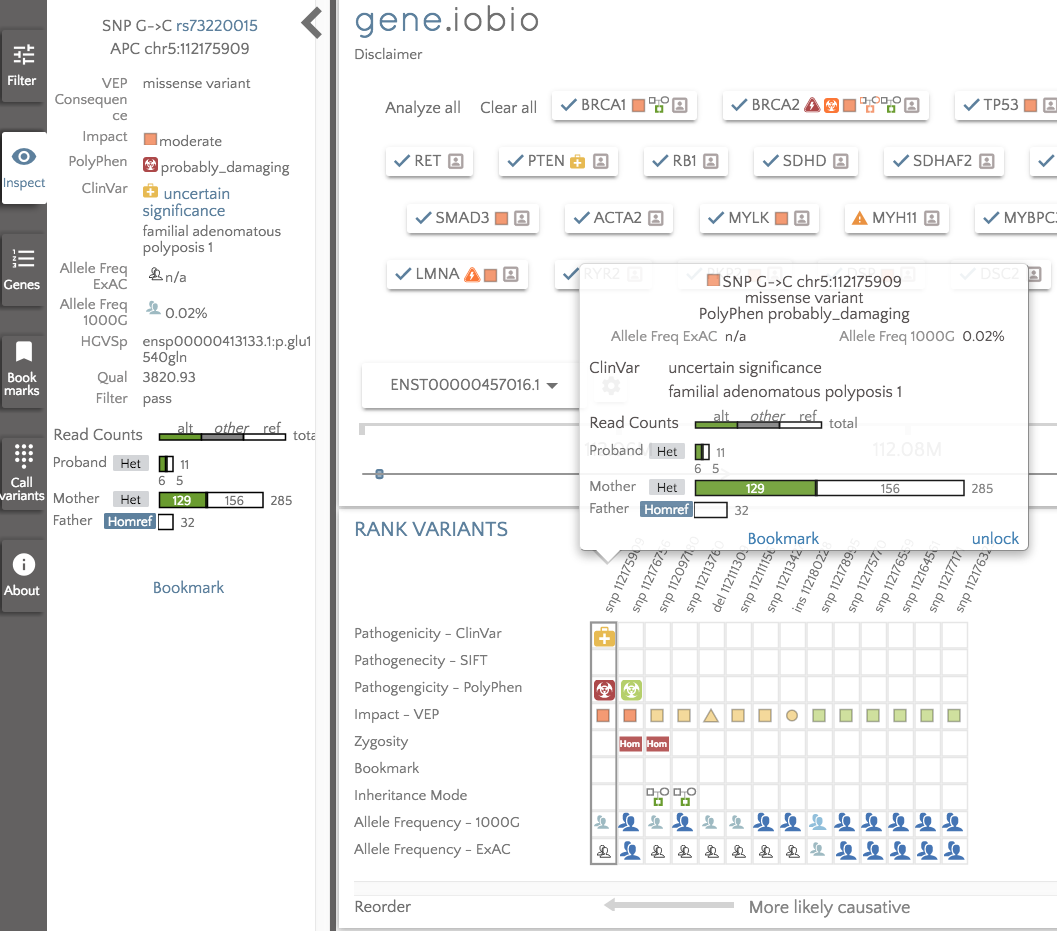
Being able to quickly analyze the ACMG56 genes gives a quick and easy way to analyze the proband in a set of genes known to be implicated in a number of diseases and have potentially actionable outcomes. This is an extremely powerful feature which we really hope the community finds useful.



