The gene.iobio version 2 blog series marches on with a discussion of multi-gene analysis. We think this is a major advancement and takes gene.iobio from a cool way of visualizing integrated data in a single gene, to a powerful method of performing analysis across sets of relevant genes. A following post in the series will look at generating gene lists based on phenotype using Phenolyzer, which really helps direct analysis.
To demonstrate functionality, we will look at the RAI1 usecase. Here we see a familiar view of the Platinum trio data, which we can interrogate as we see fit. But what if we are interested in looking at variants in a different gene? We can add another gene, say AIRE, using the gene entry box at the top of the screen. Now we see both genes have a badge below the gene entry box, with a raft of glyphs in them, as shown below:
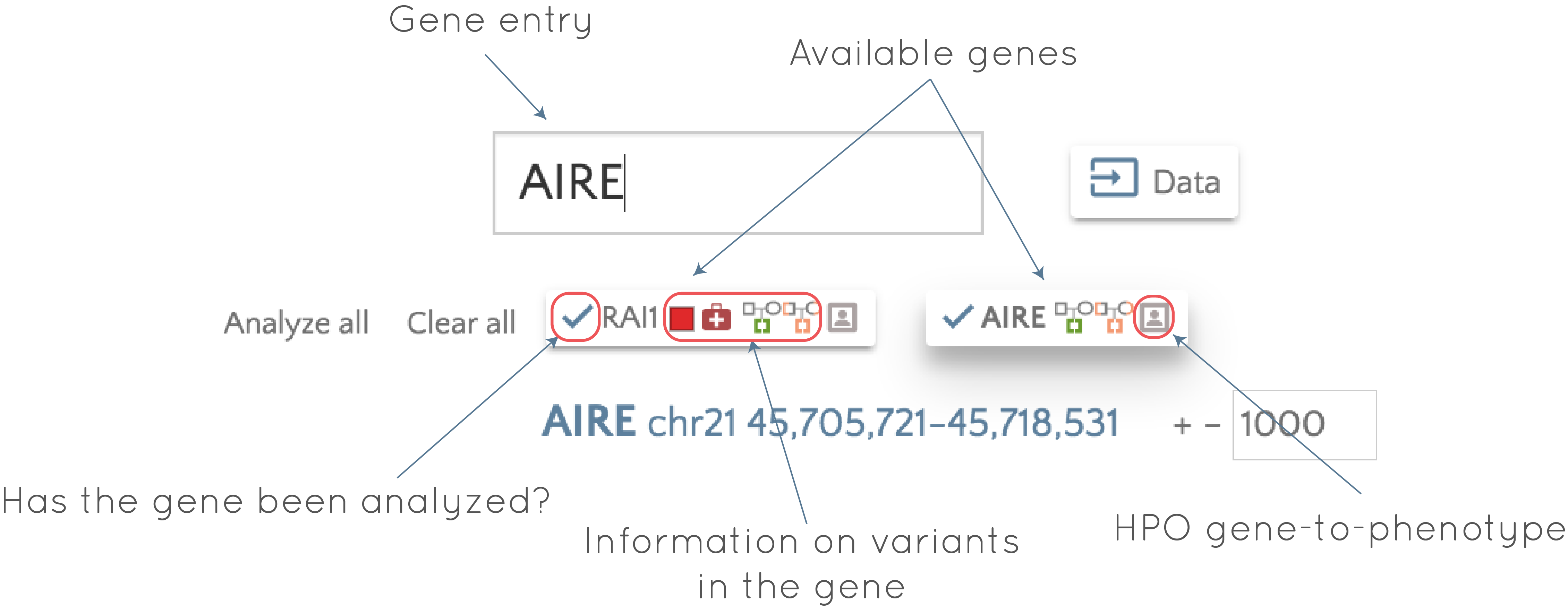
So what does all this mean? Here are some details about these gene badges and how to use them:
- Clicking on a gene badge will take you to that gene,
- If any interesting variants are present in the gene, the glyphs indicate what. In the RAI1 gene, they indicate there is a least one variant (each glyph can be referring to a different variant) in the gene that is:
- a high impact variant, as predicted by VEP (
 ),
), - a pathogenic variant in ClinVar (
 ),
), - a de novo variant (
 ),
), - a recessive variant (
 ).
). - If the checkmark (
 ) is present at the left of the gene badge, this indicates that the gene has been analyzed. The other glyphs will only be present if the gene has been analyzed, so this checkmark can differentiate between the case where the gene has been analyzed, but there are no interesting variants in the gene (
) is present at the left of the gene badge, this indicates that the gene has been analyzed. The other glyphs will only be present if the gene has been analyzed, so this checkmark can differentiate between the case where the gene has been analyzed, but there are no interesting variants in the gene ( ), or that analysis on the gene has yet to be performed (
), or that analysis on the gene has yet to be performed ( ), so we need to analyze the gene to determine if interesting variants are present.
), so we need to analyze the gene to determine if interesting variants are present. - Hovering over the HPO gene-to-phenotype glyph (
 ), when present, provides information on gene associated phenotypes. For example, these are the phenotypes associated with the RAI gene:
), when present, provides information on gene associated phenotypes. For example, these are the phenotypes associated with the RAI gene:

These gene badges make it simple to jump around a set of genes, analyzing data and identifying potentially interesting variants within them. An important question you might ask is how do we define a list of genes to look at? We can add each gene one by one, but we might want to look at a set of genes that we know are associated with a disease, or are the output of gene prioritization software. The next but one post will discuss using Phenolyzer to generate a gene list, but here we will touch on two simple methods.
Import a gene list
In the left panel, we can click the 'Genes' button to bring up a menu that lets us define gene lists. The 'Import Gene set' dropdown provides a space where we can manually import or paste a comma separated list of genes. Pressing 'Done' will prompt gene.iobio to add all defined genes as badges in the top panel so we can start working with them.
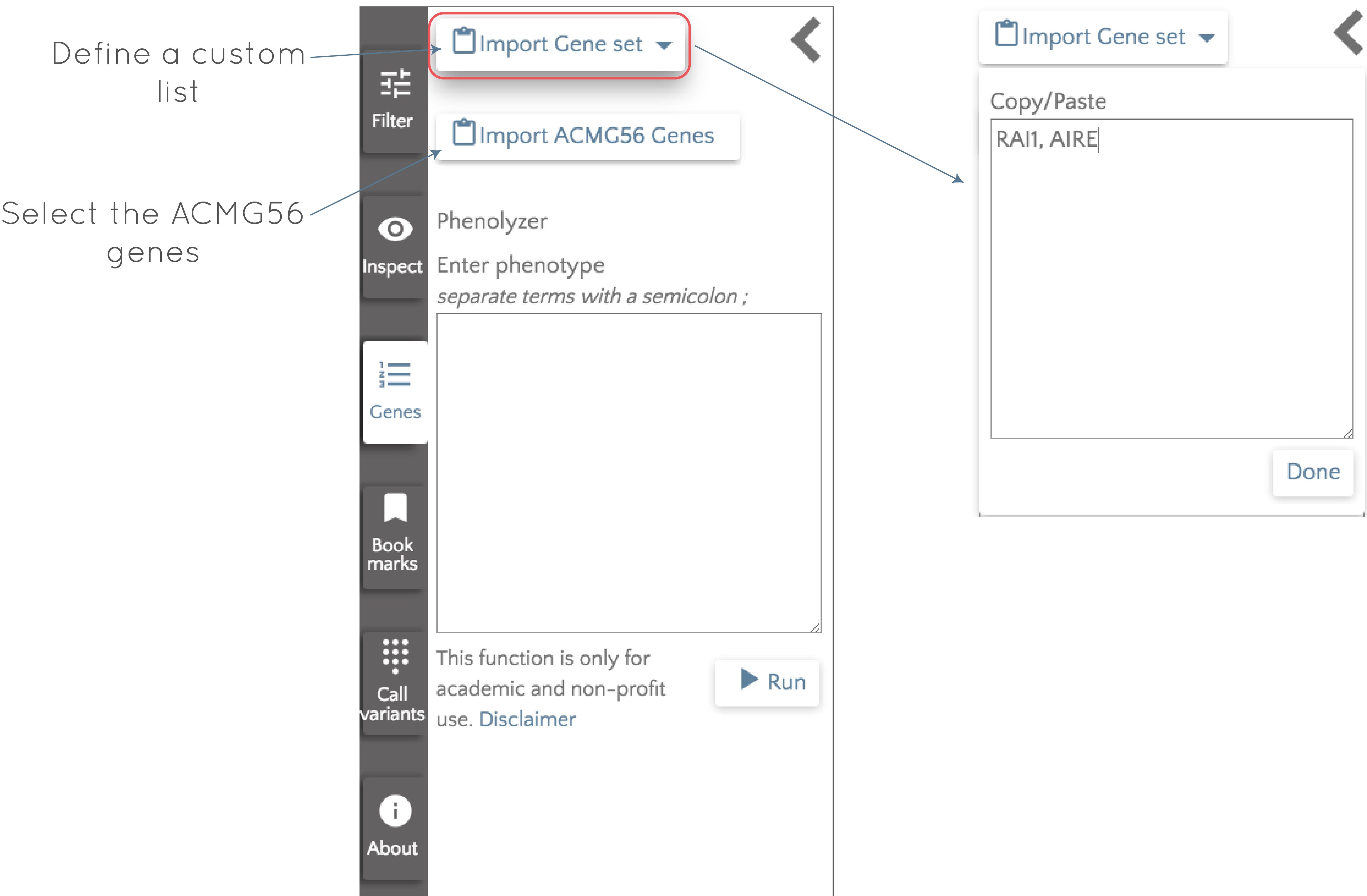
Analyze the ACMG56 genes
The American College of Medical Genetics and Genomics (ACMG) has defined a list of 56 genes that have been implicated predominantly in cancer and cardiovascular disease risk. It can be good practice to look for any known pathogenic variants in these genes, so you can add them by clicking the 'Import ACMG56 Genes' button. By clicking this, all 56 genes will be added to the top panel and be ready for analysis. The next post in the series will give a brief discussion of ths feature.
Analyze all genes
Once a set of genes has been defined, the next step is usually to see if any of them harbour potentially interesting variants. Preferably, we would like to achieve this without having to click through every gene individiually. Luckily, we are not forced to do that! Instead, we can just click 'Analyze all' to the left of the gene badges and each gene will be analyzed in turn; the gene being analyzed has a working symbol ( ) to identify which gene is being processed. As each gene is analyzed the check mark (
) to identify which gene is being processed. As each gene is analyzed the check mark ( ) appears along with any glyphs describing contained variants. Once complete, the glyphs appearing in each gene badge direct us to where interesting variants lie (note that you can click on an analyzed gene to investigate, while gene.iobio continues to work through the rest of the genes).
) appears along with any glyphs describing contained variants. Once complete, the glyphs appearing in each gene badge direct us to where interesting variants lie (note that you can click on an analyzed gene to investigate, while gene.iobio continues to work through the rest of the genes).
As always, contact us with any questions or comments, and stay tuned for the next two posts describing using the ACMG56 genes and the use of Phenolyzer to turn your phenotype into a prioritized gene list.



