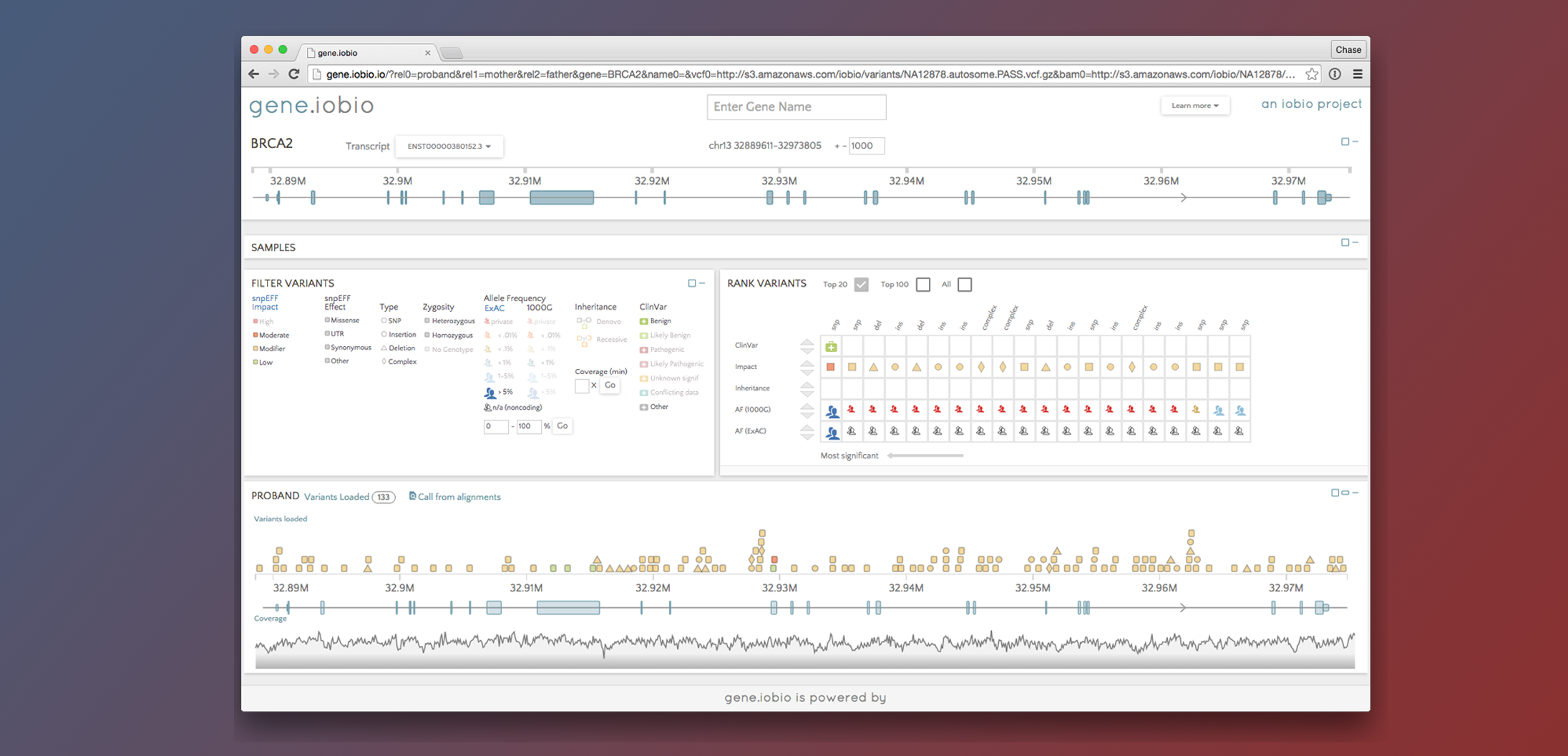
We’ve been working on a new real-time interactive iobio analysis tool for the last few months and today are excited to officially release gene.iobio. Gene.iobio is designed to help medical and clinical researchers hunt for disease-causing genetic variants through a combination of real-time genomic data analysis and intuitive visualization. The quickest way to get an idea of the features and functionality is to watch the intro screencast, quite professionally voiced over by our own Alistair Ward!
Gene.iobio integrates many tools and data sources into a single intuitive and user-friendly web interface. The app allows a researcher to rapidly examine genetic variants in a patient sample according to predicted functional impact (determined via snpEff), and in the context of the pattern of inheritance from the parents. It supports the visualization of sequence alignments in the samples to assess coverage, and even to call potentially missed variants in real time. The app also pulls in data from 1000 Genomes Project and ExAC to see if a given variant in the patient is common or rare in the general population. Additional touches include the ability to reanalyze variant impact from non-canonical transcripts / gene models.
Importantly, gene.iobio (just like all iobio apps) does not require any data upload. You can simply select the data files and the gene of interest, and the app fetches all the data in that gene in a couple of seconds.
On the gene.iobio website you can find additional screencasts and descriptions of several practical use cases.
We are very excited about this app and hope to continue to improve it. If you have any questions or feedback leave a comment below or email us at iobioproject@gmail.com