gene.iobio 3.0 is live!
The Marth lab is thrilled to announce the latest version of gene.iobio, our biggest upgrade yet. The features within this release were incorporated largely based on feedback from clinicians and diagnostic pathologists. We hope these improvements will further optimize how users interact with gene.iobio.
In addition to powerful new features and improvements, we have also converted gene.iobio to Vue.js, a Javascript framework that allows us to continue adding new features and improve user interfaces. As you’ll notice, this new framework has allowed us to dramatically redesign and simplify the user interface.
Here’s a quick summary of the new gene.iobio interface.
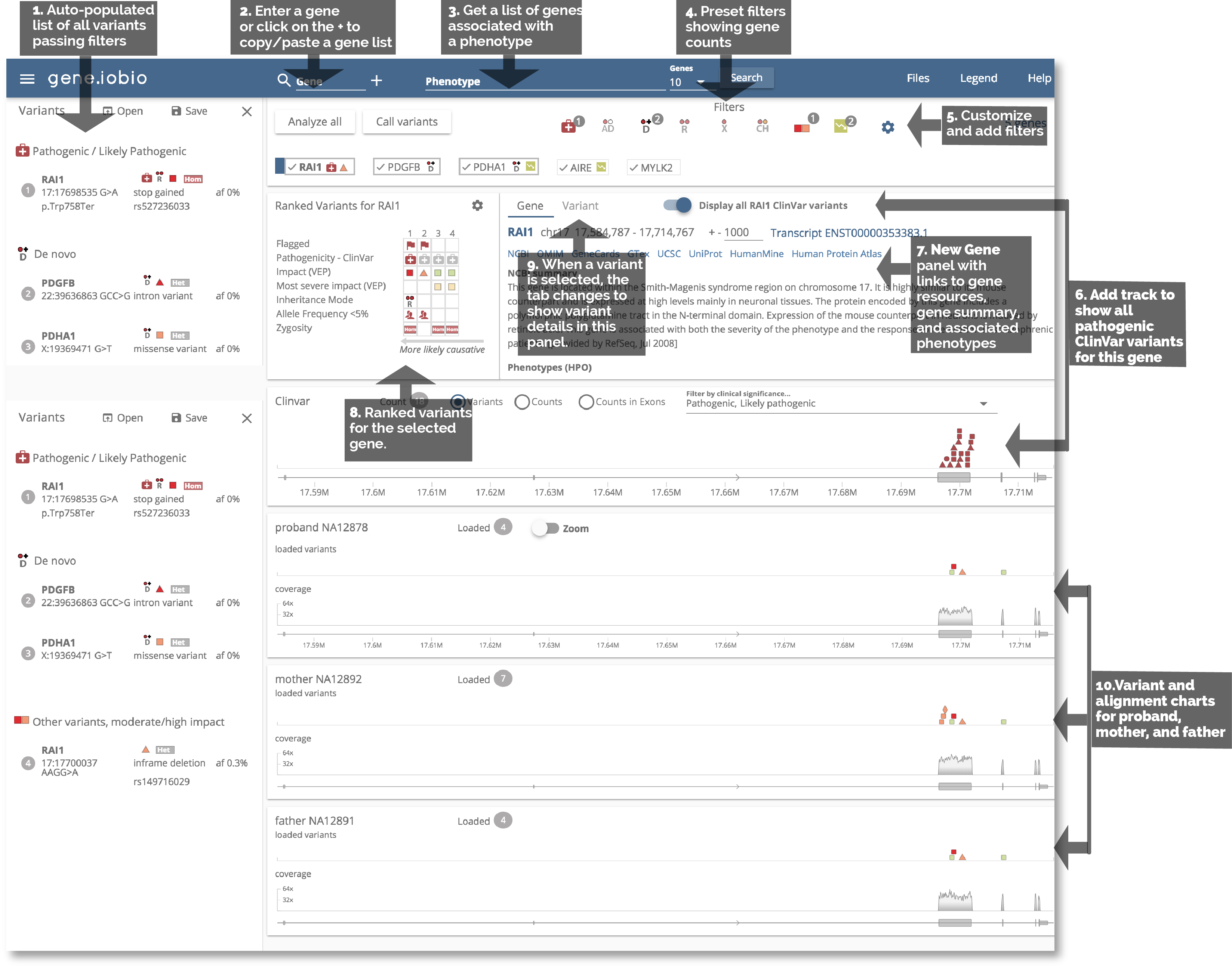
1. Automatic analysis and preset filters
After loading a gene or gene list, analysis is started automatically and any variants that pass the preset filters are automatically populated in the left side panel. Quickly jump between these variants by simply clicking on their entries.
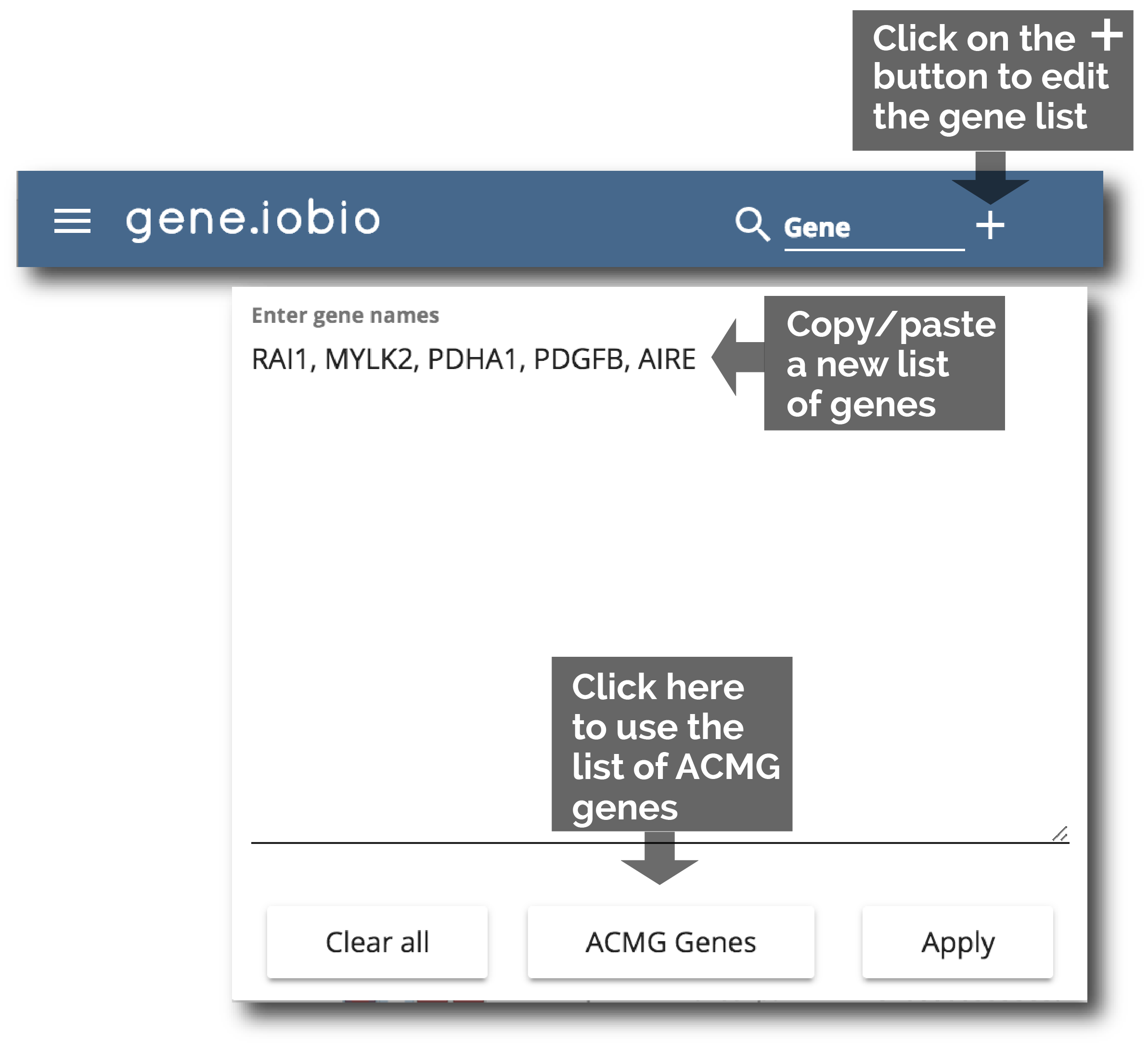
2. (and 3.) Rapid gene and phenotype input
Easily manage your gene list by simply clicking the + button in the gene bar. A drop-down allows you to paste a new gene list or load the ACMG genes.
Phenotype-driven gene input is now built directly into the navigation bar. Just start typing a phenotype term, select your specific term from the dropdown and genes associated with that phenotype will automatically be loaded. (Thanks Phenolyzer!)

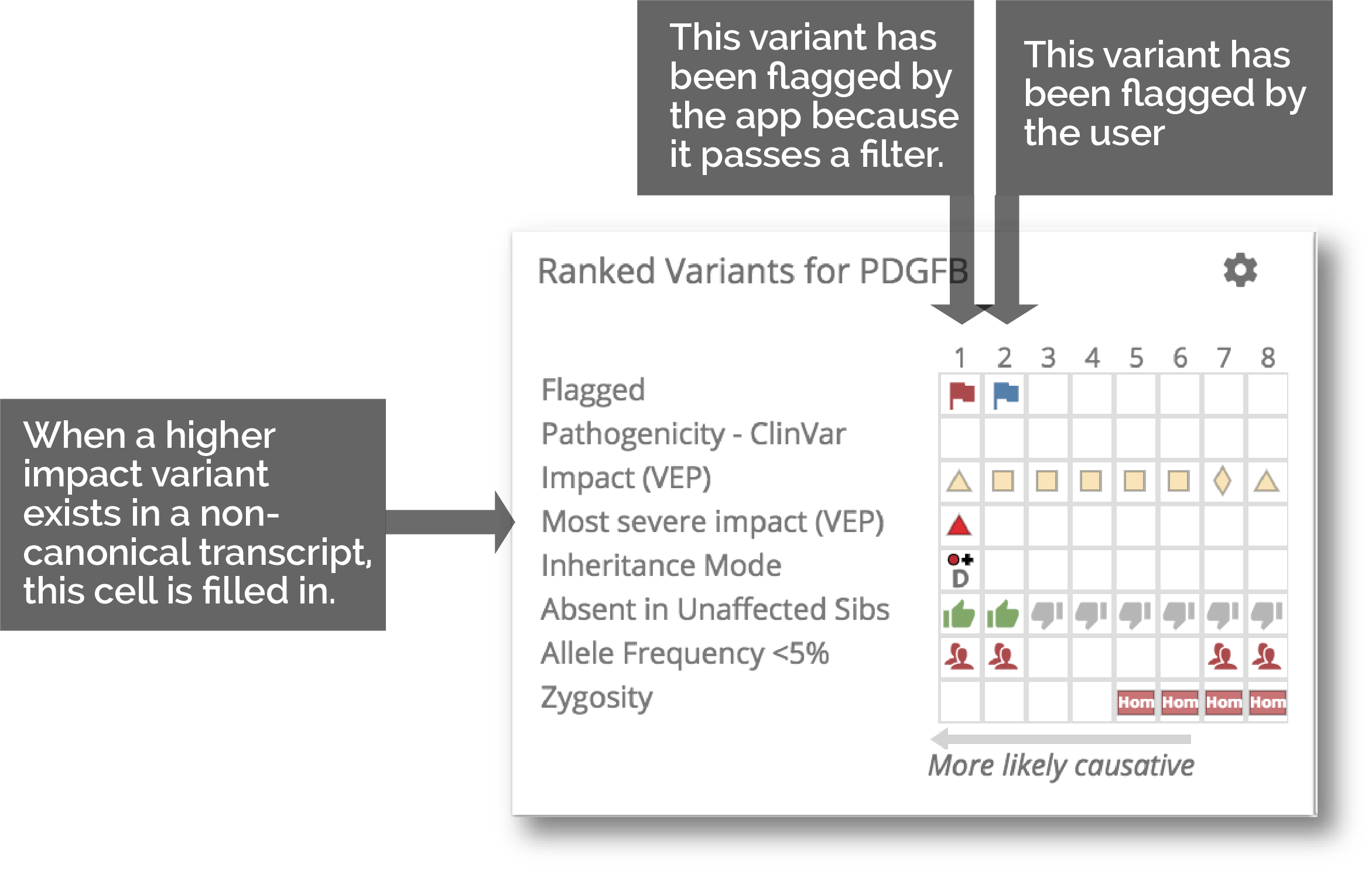
4. Automatic flagging of potentially interesting variants
As genes are analyzed in real-time, variants that meet preset filter criteria are flagged into their respective filter badges. This allows you to quickly see how many variants meet these criteria in your gene list, without having to click through every gene. Simply click on the badge to see all the genes that pass that filter.

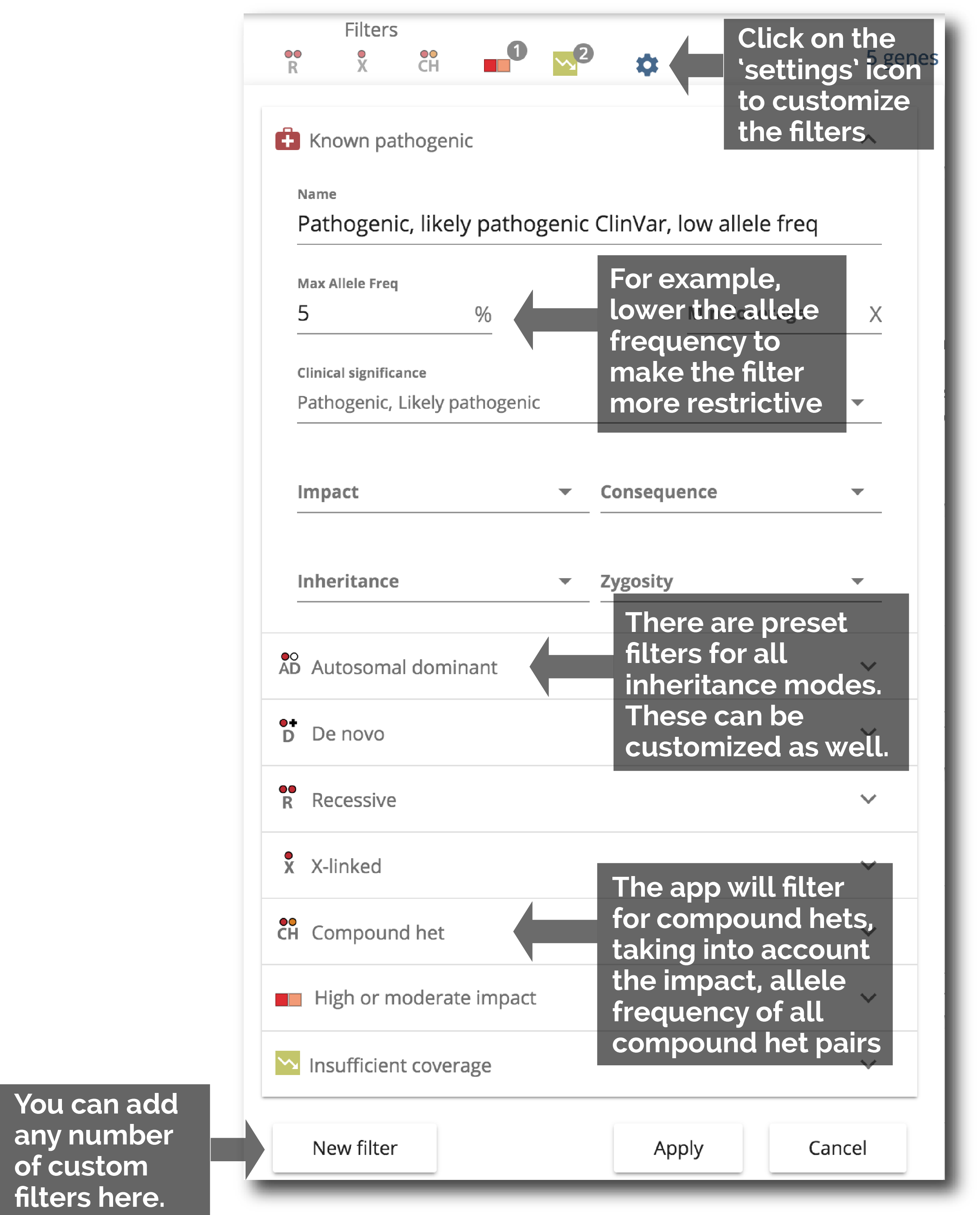
5. Further customize filters
Customize the preset filters and create your own filters. When filters change, all variants are re-analyzed and flags are updated.
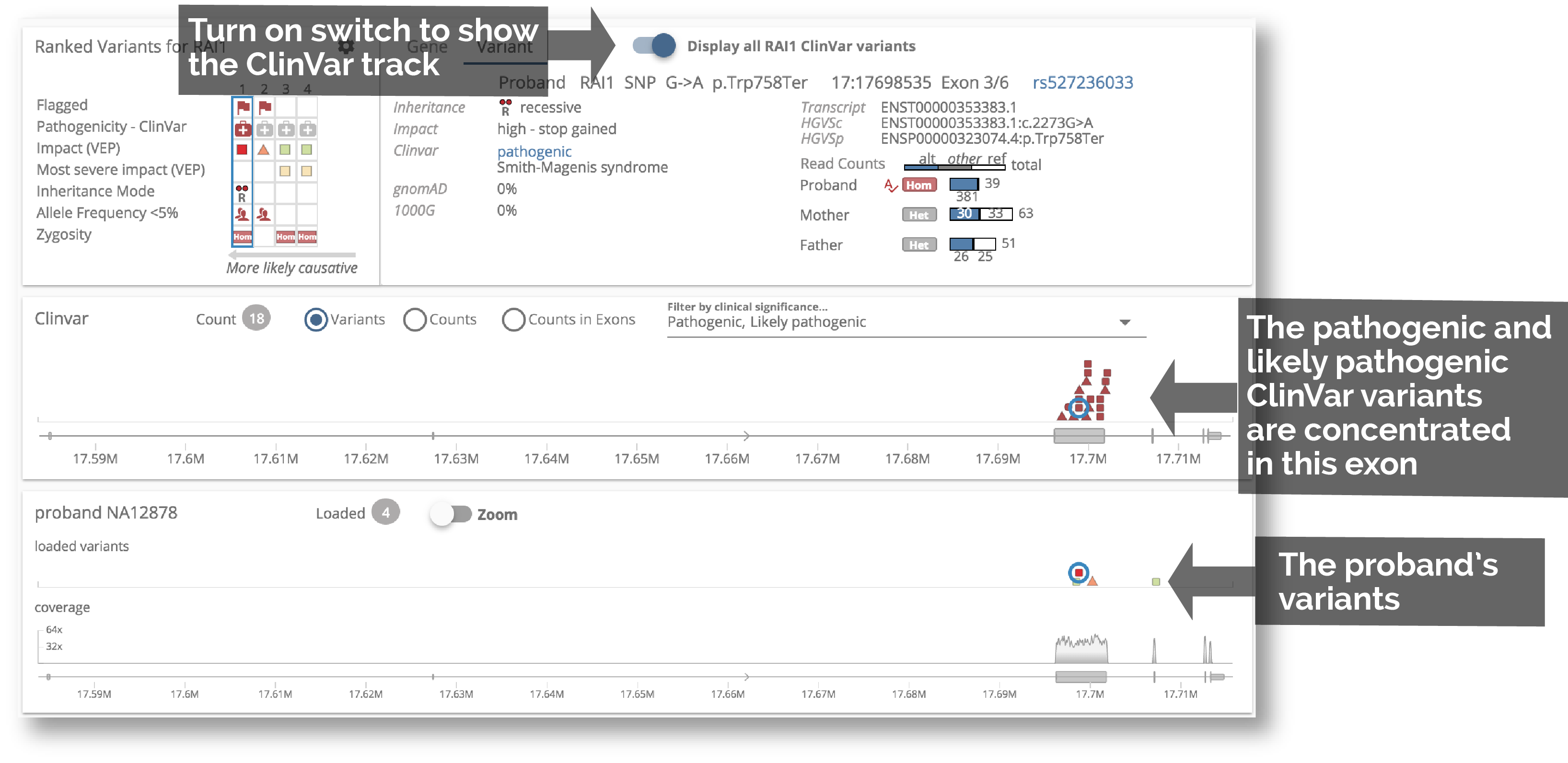
6. Visually compare to ClinVar variants
Have an interesting variant, but not sure about pathogenicity? Compare your variants to variants in ClinVar, easily determining whether they fall within the same gene region as known pathogenic variants.
7. Quickly launch gene resources
A large number of considerations must be made when determining variant pathogenicity as variant/gene resources continue to grow. Now you can quickly get more information about a particular gene by clicking the resource links under the gene name.

8. Ranked Variants table
We have kept the ranked variants table front and center to easily navigate between the the highest ranked variants in a gene.
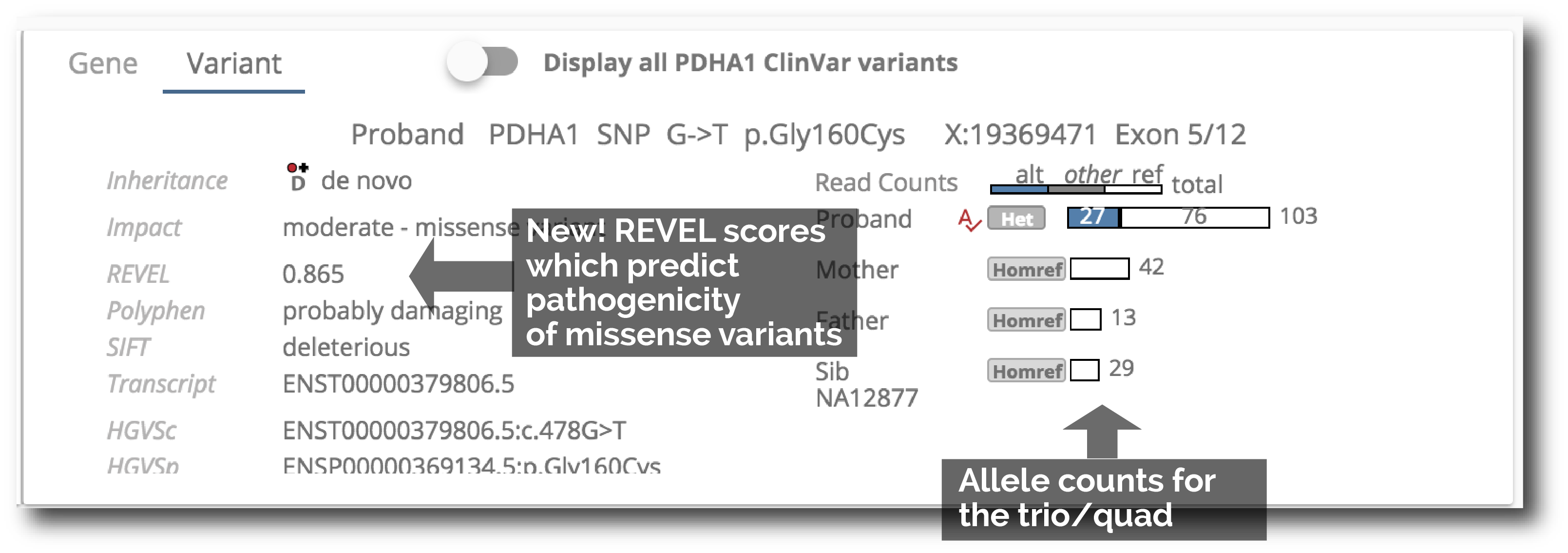
9. Comprehensive variant details in a single pane
The gene information pane switches over to a variant information pane when a variant is clicked. This variant pane gives you comprehensive variant information, including REVEL scores for missense variant pathogenicity prediction.
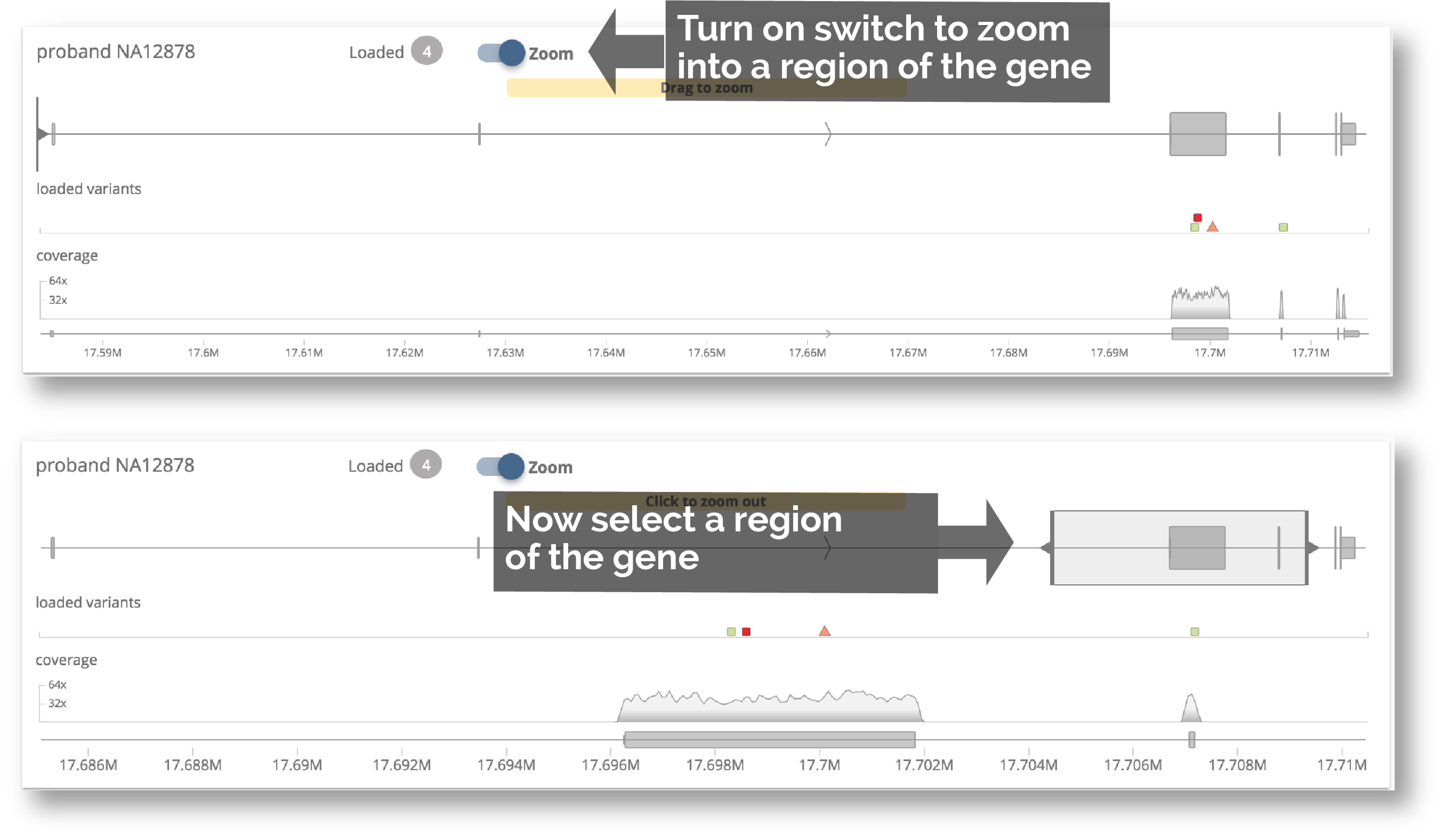
10. Variant and alignment charts for Trio
We have made it easier to zoom into a region of the gene. Also, now that the large tooltip has been replaced with the Variant Detail panel, the charts are no longer obscured. We have also tried to save vertical space so that these charts are visible without scrolling.